宏基因组学及宏转录组学分析工具MOCAT2(Meta‘omic Analysis Toolkit 2)安装配置及常用使用方法
详细介绍
尽管这个工具已经暂停后续开发,但其工具功能还是挺好的,大家可以参考一下,尤其对于喜欢自定义开发流程的可以参考是流程。

MOCAT 2(Meta'omic Analysis Toolkit 2)是一个用于宏基因组和宏转录组数据分析的工具集,旨在处理和分析来自各种环境样品(如土壤、水体、肠道等)的宏基因组学和宏转录组学数据。它提供了一系列功能模块,涵盖了数据预处理、序列比对、装配、功能注释和分析等方面。
文章:
MOCAT: A Metagenomics Assembly and Gene Prediction Toolkit | PLOS ONE
官网:MOCAT2
github:GitHub - mocat2/mocat2: Latest MOCAT2 version
 ?MOCAT 2的主要特点和功能:
?MOCAT 2的主要特点和功能:
-
综合性:支持宏基因组学和宏转录组学数据的处理和分析,能够应用于不同类型的元组件(metagenome、metatranscriptome)数据。
-
数据预处理:包括质量控制、去除PCR重复、去除低质量序列、去除宿主序列等预处理步骤,以准备数据用于后续分析。
-
序列比对:能够进行序列比对到参考数据库,对元组件数据进行分类、注释等。
-
序列装配:支持元组件数据的序列装配,得到组装得到的序列。
-
功能注释和分析:提供了功能注释和分类分析模块,能够进行基于注释的功能分析,如基因功能注释、基因家族分析等。
-
并行计算:支持多线程并行计算,能够加速数据处理和分析的速度。
-
灵活性:提供了多种配置选项和参数,可根据不同的实验设计和数据类型进行定制化处理和分析。
-
支持多种数据格式:能够处理和分析常见的测序数据格式,如FASTQ、FASTA等。
MOCAT 2的使用流程:
MOCAT 2的使用流程包括数据准备、选择合适的模块和参数、运行分析、结果解释和分析等步骤。用户可以根据实验设计和数据类型选择合适的模块和参数进行分析,并根据分析结果进行后续的生物信息学分析或实验设计。
官方文档和资源:
MOCAT 2提供了详细的官方文档和使用指南,其中包括安装指南、使用教程、参数说明等,可在官方网站或GitHub页面获取相关信息和支持:
MOCAT2(Meta'omic Analysis Toolkit 2)是用于宏基因组和宏转录组数据分析的工具,提供了一系列功能用于质量控制、序列比对、装配、注释等。以下是MOCAT2的基本使用方法和分析流程:
安装 MOCAT2:
MOCAT2可以从其官方网站或GitHub页面获取源代码,并且在Linux环境下进行编译安装。可以参考官方文档提供的安装指南进行安装:MOCAT2 GitHub
1. 安装依赖项
在开始安装MOCAT2之前,需要确保系统中已安装以下依赖项:
- Python 2.7 或更高版本
- C++ 编译器(如GCC)
- Perl 5 或更高版本
- Perl模块:DB_File、Math::Round、List::Util、Digest::MD5
- 某些功能可能需要安装其他外部工具,如Bowtie2、BLAST等
2. 下载源代码
可以从MOCAT2的GitHub页面获取源代码。在终端中运行以下命令来克隆MOCAT2的代码库:
git clone https://github.com/mocat2/mocat2.git?进入安装目录,运行设置脚本
#
cd mocat2/stable/2.1.3
./setup.MOCAT2.pl
#或
perl ./setup.MOCAT2.pl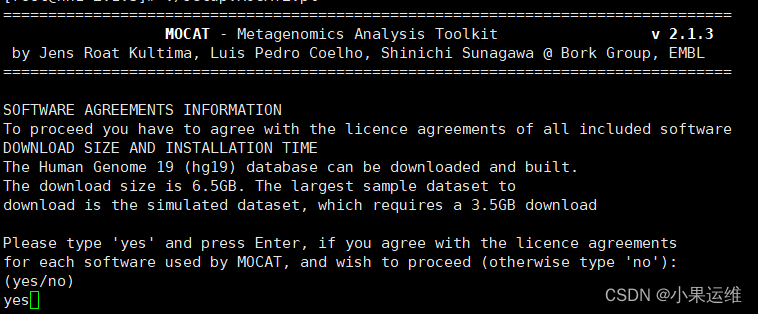 ?要不要下载扩展数据库或要不要下载文章数据集,我这里都选择了no,因为感觉用不上。大家可根据需求下载
?要不要下载扩展数据库或要不要下载文章数据集,我这里都选择了no,因为感觉用不上。大家可根据需求下载

MOCAT2的一些常见模块及其输出结果文件的内容展示和介绍:
1. mocat_preprocessing 模块:
- 输出文件:
clean_reads_1.fastq、clean_reads_2.fastq: 经过质量控制和预处理后的测序数据。summary_statistics.txt: 包含关于质量控制步骤的统计信息,如序列数目、质量分数统计等。
2. mocat_assembly 模块:
- 输出文件:
contigs.fasta: 组装得到的contigs序列。assembly_stats.txt: 包含有关组装质量和性能的统计信息,如N50、最大/最小contig长度等。
3. mocat_analysis 模块:
- 输出文件:
blast_results.txt:包含BLAST注释的结果,显示序列与参考数据库的相似性。gene_catalog.fasta:根据比对结果生成的基因目录序列。functional_annotation.txt:功能注释的结果文件,包括基因或序列的功能描述、KEGG或COG注释等信息。classification_results.txt:分类结果,显示序列或基因的分类信息,如菌株、属、门水平的分类等。
4. mocat_metaquant 模块(可选,用于定量分析):
- 输出文件:
gene_abundance_table.txt:基因丰度表,显示每个基因在样本中的丰度估算。transcript_abundance_table.txt:转录本丰度表,显示转录本在样本中的丰度估算。- 其他可能包含样本丰度信息的文件。
注意事项:
- 每个模块生成的输出文件格式和内容可能会因应用不同参数和实验设计而有所不同。
- 结果文件中包含的信息可以帮助研究人员了解数据质量、序列注释信息、组装质量和功能注释等方面的信息。
- 输出文件中的数据通常以文本或FASTA等格式呈现,可以使用文本编辑器或专业的生物信息学软件进行查看和进一步分析。
MOCAT2 使用流程:
数据准备:
- 获得宏基因组/宏转录组测序数据(FASTQ格式)。
- 准备参考数据库,如基因组数据库或功能注释数据库。
运行 MOCAT2:
MOCAT2的主要模块和使用示例命令如下:
mocat_preprocessing:进行质量控制和预处理。
mocat_preprocessing -t 4 -o output_directory --input-files reads_1.fastq,reads_2.fastqmocat_assembly:执行序列组装。
mocat_assembly -t 4 -o output_directory --input-files reads_1.fastq,reads_2.fastqmocat_analysis:进行功能注释和分类分析。
mocat_analysis -t 4 -o output_directory --input-files assembly.fa这里的 -t 选项用于指定线程数,-o 用于指定输出目录,--input-files 用于指定输入文件。
结果解释和分析:
MOCAT2生成的输出文件包括装配得到的序列、注释结果、分类信息等。可以使用其他工具或分析流程进一步解释和分析这些结果。
示例代码:
以下是一个使用MOCAT2的简单Shell脚本示例,演示了一个简单的分析流程:
# 质量控制和预处理
mocat_preprocessing -t 4 -o preprocessing_output --input-files reads_1.fastq,reads_2.fastq
# 序列组装
mocat_assembly -t 4 -o assembly_output --input-files preprocessing_output/clean_reads_1.fastq,preprocessing_output/clean_reads_2.fastq
# 功能注释和分类分析
mocat_analysis -t 4 -o analysis_output --input-files assembly_output/contigs.fasta
注意事项:
- MOCAT2提供了丰富的功能和模块,具体的使用方法和参数设置需要根据数据类型和实验设计进行调整。
- 分析过程可能需要较长的时间和较大的计算资源,特别是对于大规模的宏基因组/宏转录组数据。
- 根据数据类型和分析需求,可能需要进一步的后续分析和解释。
MOCAT.pl全参数帮助信息
MOCAT.pl --help
===============================================================================
MOCAT - Metagenomics Analysis Toolkit v2.1.3
by Jens Roat Kultima, Luis Pedro Coelho, Shinichi Sunagawa @ Bork Group, EMBL
===============================================================================
Full manual & FAQ: MOCAT.pl -man
How to cite MOCAT: MOCAT.pl -cite
Have you tried the wrapper runMOCAT.sh? Try it!
Usage: MOCAT.pl -sf|sample_file 'FILE' [Pipeline, Statistics, & Additional Options]
'FILE'
Contains the list of folder names (sample names), one per line,
in which the raw sample data is located
Examples
Process, Assemble, Revise Assembly, Predict Genes, cluster genes into gene catalog, annotate gene catalog, profile against gene catalog
MOCAT.pl -sf my.samples -rtf
MOCAT.pl -sf my.samples -a
MOCAT.pl -sf my.samples -gp assembly
MOCAT.pl -sf my.samples -make_gene_catalog -assembly_type assembly
MOCAT.pl -sf my.samples -annotate_gene_catalog
MOCAT.pl -sf my.samples -s my.samples.padded -identity 95
MOCAT.pl -sf my.samples -f my.samples.padded -identity 95
MOCAT.pl -sf my.samples -p my.samples.padded -identity 95 -mode functional
Assemble and predict genes: MOCAT.pl -sf my.samples -rtf
(no screen) MOCAT.pl -sf my.samples -a
MOCAT.pl -sf my.samples -gp assembly
fetch marker genes: MOCAT.pl -sf my.samples -fmg assembly
MOCAT.pl -sf my.samples -ss
Assemble and predict genes: MOCAT.pl -sf my.samples -rtf
(DB screen) MOCAT.pl -sf my.samples -s hg19 -screened_files -identity 90
MOCAT.pl -sf my.samples -a -r hg19
MOCAT.pl -sf my.samples -gp assembly -r hg19
MOCAT.pl -sf my.samples -ss
Assemble and predict genes: MOCAT.pl -sf my.samples -rtf
(remove eg. adapters MOCAT.pl -sf my.samples -sff adapters.fa -screened_files
and then DB screen) MOCAT.pl -sf my.samples -bwa hg19 -r adapters.fa -screened_files
MOCAT.pl -sf my.samples -a -r screened.adapters.fa.on.hg19
MOCAT.pl -sf my.samples -gp assembly -r screened.adapters.fa.on.hg19
MOCAT.pl -sf my.samples -ss
Pipeline Options
-r|reads ['reads.processed', 'DATABASE' or 'FASTA FILE']
Required for all pipeline options, except rtf|read_trim_filter
Specify whether processing trim & filtered, or screened reads.
A default value to this setting can also be specified in config file
-e|extracted
Optional for all pipeline options, except rtf|read_trim_filter, see full manual
-rtf|read_trim_filter
performs trimming and filtering of reads
-a|assembly
Performs assembly of reads
-ar|assembly_revision
Further improves assemblies
-gp|gene_prediction ['assembly', 'assembly.revised']
Predicts protein coding genes on assemblies
-fmg|fetch_mg ['assembly', 'assembly.revised']
Extracts marker genes among the predicted genes
-soap|bwa ['DB1 DB2 ...',s,c,f,r]
Screen, extract and map reads against a reference databse (hg19 is provided) or (s)acftigs,
(c)ontigs, sca(f)folds from an assembly, or scaftigs from a (r)evised assembly.
This mapping step uses SOAPaligner2 (soap) or BWA (bwa).
Additional options:
-screened_files : If set, screened read files are generated, these are reads not matching the DB
-extracted_files : If set, extracted read files are generated, these are reads matching the DB
-use_mem : If set, copies the DB into memory for faster loading
-sff|screen_fastafile 'FASTA FILE'
Same as 's|screen' above, but uses USearch, rather than SOAPaligner2.
-fsoap ['DB1 DB2 ...',s,c,f,r]
Filter screened reads, (s)caftigs, (c)ontigs, sca(f)folds or (r)evised assembly scaftigs
at higher %ID and length cutoff. This step has to be run before calculating profiles if the option soap was used
Additional options:
-shm : If set, faster, but saves data for the filtering step in /dev/shm/<USER>
-psoap|pbwa ['DB1 DB2 ...',s,c,f,r] -m|mode [gene, NCBI, mOTU, functional] -o [OUTPUT FOLDER]
Generate gene, mOTU, NCBI or functional profiles on filtered reads,
(s)caftigs, (c)ontigs, sca(f)folds or (r)evised assembly scaftigs.
If -mode is set to either NCBI or mOTU, it is expected that the
reads have been correctly mapped to the corresponding databases.
Specify psoap if you used the command 'soap' previously, and 'pbwa' if you used 'bwa'.
Additional options:
-no_horizontal : No not calculate horizontal gene & functional coverages
-verbose : Prints extra information about status of profiling steps
-shm : Faster, but saves 2-5 GB of data for the profiling step in /dev/shm/<USER>
-uniq : Specify this flag if you find duplicated row names
(e.g. if you have mapped to a DB where the same reference appears multiple times)
Available modules
These are installed in the folder /nfs/data/Downloads/mocat2/stable/2.1.3/mod
Each module requires a NAME.sh and NAME.cfg file inside the NAME folder
-annotate_gene_catalog [leave empty for using sample file generated catalog or enter full path to catalog; use amino acid sequence file]
Required options:
-blasttype [should be "blastp" normally for amino acid sequences, but can be set to "blastx"]
-make_gene_catalog [samples specifed in sample file will be used ot generate catalog]
Required options:
-assembly_type [asembly or assembly.revised]
Statistics Options
-sfq|stats_fastqc
Produces statistics for each lane with raw reads using the FastQC toolkit
-ss|sample_status
Prints a simple view how the processing status of each sample,
and stores this in <sample_file>.status
Additional Options
-cfg|config [file]
Specify another config file than MOCAT.cfg
-x|no_execute
Only create job scripts, but don't execute them
-nt|no_temp
Overrides any specified temp folders config file
-cpus [integer]
Not recommended, but specifies a fixed number of cores for each job,
please read the full manual using MOCAT.pl -man
-host [hostname]
Runs the jobs on a different host machine
-identity [integer]
Overrides any percentage cutoff setting in cfg file
-length [integer]
Overrides any length cutoff setting in cfg file
-memory XGB
If queuing system is SGE or LSF, it will require XGB of RAM for the job
This can also be set with the respective memory options by adding these
to the param fields in the config file
-config A=b C=d
Overrides setting A from the config file with b, etc
本文来自互联网用户投稿,该文观点仅代表作者本人,不代表本站立场。本站仅提供信息存储空间服务,不拥有所有权,不承担相关法律责任。 如若内容造成侵权/违法违规/事实不符,请联系我的编程经验分享网邮箱:chenni525@qq.com进行投诉反馈,一经查实,立即删除!
- Python教程
- 深入理解 MySQL 中的 HAVING 关键字和聚合函数
- Qt之QChar编码(1)
- MyBatis入门基础篇
- 用Python脚本实现FFmpeg批量转换
- 【产品】AI领域专业术语学习笔记
- 【python基础】不懂什么是面向对象?没关系一篇文章教会你
- 【ThreeJS入门——】WEB 3D可视化技术——threejs
- 第二百八十五回
- 【超详细教程】GoogleTest CMake直接构建(无需安装,手把手教程)
- 世界上真的存在比苹果品质还好的笔记本电脑吗?
- 使用Python创建和操作序列
- 某出版社薪酬管理项目成功案例
- 第11章 1 文件及IO操作
- 我对前端/互联网发展的一些看法
