Cell 文章图复现
发布时间:2024年01月04日
多组差异火山图复现
参考文章: A Spatiotemporal Organ-Wide Gene Expression and Cell Atlas of the Developing Human Heart Figure 2. H
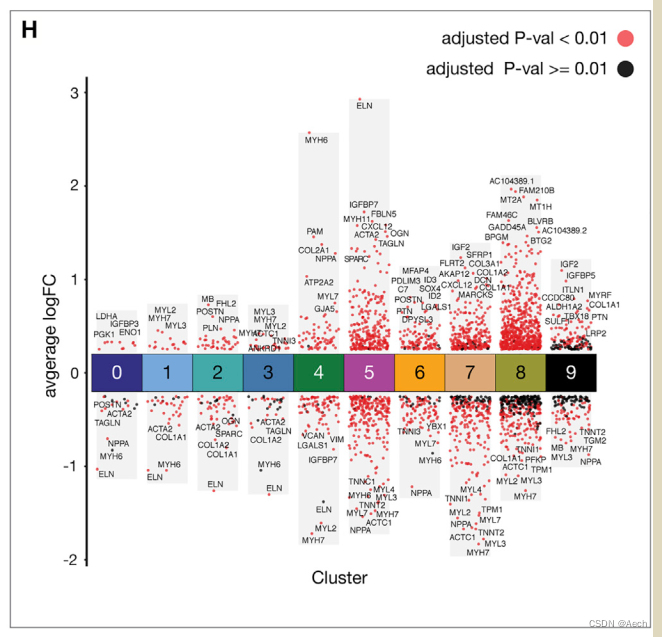
图里主要是单细胞数据不同cluster之间的差异火山图, 所以说白了就是散点图和柱状图的结合, 散点图用差异基因绘制, 柱状图利用logFC最大最小值绘制就完了.
加载包
> library(tidyverse)
> library(ggplot2)
> library(ggpubr)
> library(RColorBrewer)
> library(openxlsx)
> library(ggsci)
> library(ggrepel)
> # Create color parameters
> qual_col_pals = brewer.pal.info[brewer.pal.info$category == 'qual',]
> col_vector = unlist(mapply(brewer.pal, qual_col_pals$maxcolors, rownames(qual_col_pals)))
>
读取数据
> deg <- read.csv("./Differentially_Expressed_Markers_Each_Cluster.csv", header = T)
> deg$cluster <- as.factor(deg$cluster)
> head(deg)
X p_val avg_log2FC pct.1 pct.2 p_val_adj cluster gene
1 1 0 2.558924 0.982 0.289 0 0 DEFB1
2 2 0 2.365316 0.963 0.220 0 0 HMGCS2
3 3 0 2.317304 0.991 0.513 0 0 ATP1B1
4 4 0 2.207154 0.963 0.231 0 0 AC015522.1
5 5 0 2.153153 0.912 0.244 0 0 HSD11B2
6 6 0 2.125726 0.811 0.209 0 0 PAPPA2
> deg <- deg %>% dplyr::filter(p_val_adj < 0.05) %>%
+ dplyr::filter(abs(avg_log2FC) > 0.75) %>%
+ dplyr::select(avg_log2FC, p_val_adj, cluster, gene) # filter and tidy the matrix
>
添加一些注释信息, 例如legend, 上下调, 需要显示名称的基因等
> deg <- deg %>%
+ mutate(label = ifelse(p_val_adj < 0.01, "adjusted P-val < 0.01", "adjusted P-val >= 0.01")) %>%
+ mutate(Change = ifelse(avg_log2FC > 0.75, "UP", "DOWN"))
>
> bardata <- deg %>% dplyr::select(cluster, avg_log2FC ) %>%
+ group_by(cluster) %>%
+ summarise_all(list(tail = min, top = max)) #
> head(bardata)
# A tibble: 6 × 3
cluster tail top
<fct> <dbl> <dbl>
1 0 -5.61 2.56
2 1 -5.13 4.32
3 2 -5.46 2.53
4 3 -4.84 4.81
5 4 -5.60 3.97
6 5 -4.59 2.96
>
> tagedgene <- deg %>% group_by(cluster) %>%
+ slice_max(abs(avg_log2FC), n = 3)
> head(tagedgene)
# A tibble: 6 × 6
# Groups: cluster [2]
avg_log2FC p_val_adj cluster gene label Change
<dbl> <dbl> <fct> <chr> <chr> <chr>
1 -5.61 0 0 ALDOB adjusted P-val < 0.01 DOWN
2 -5.46 0 0 HSPA1A adjusted P-val < 0.01 DOWN
3 -5.09 0 0 GPX3 adjusted P-val < 0.01 DOWN
4 -5.13 0 1 DEFB1 adjusted P-val < 0.01 DOWN
5 -4.61 0 1 CRYAB adjusted P-val < 0.01 DOWN
6 -4.36 1.07e-43 1 ALDOB adjusted P-val < 0.01 DOWN
>
绘制图形
- 利用bardata绘制背景柱状图
ggplot(deg, aes(x = cluster, y = avg_log2FC ))+
geom_col(data = bardata, mapping = aes(x = cluster, y = tail),
fill = "grey", width = 0.8) +
geom_col(data = bardata, mapping = aes(x = cluster, y = top),
fill = "grey", width = 0.8)
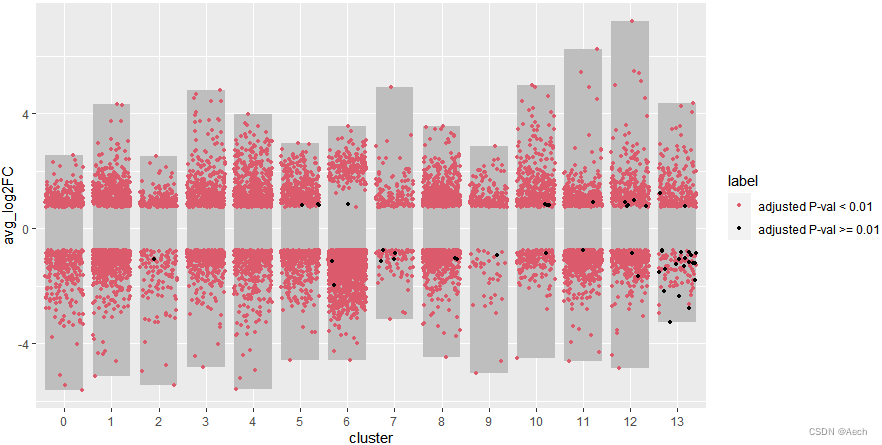
- 添加上散点图, 黑色点有点少了,
不过无所谓能看到就行
ggplot(deg, aes(x = cluster, y = avg_log2FC ))+
geom_col(data = bardata, mapping = aes(x = cluster, y = tail),
fill = "grey", width = 0.8) +
geom_col(data = bardata, mapping = aes(x = cluster, y = top),
fill = "grey", width = 0.8) +
geom_jitter(aes(color = label), size = 1,
position = position_jitter(seed = 0328)) +
scale_color_manual(values = c("#db5a6b", "black"))
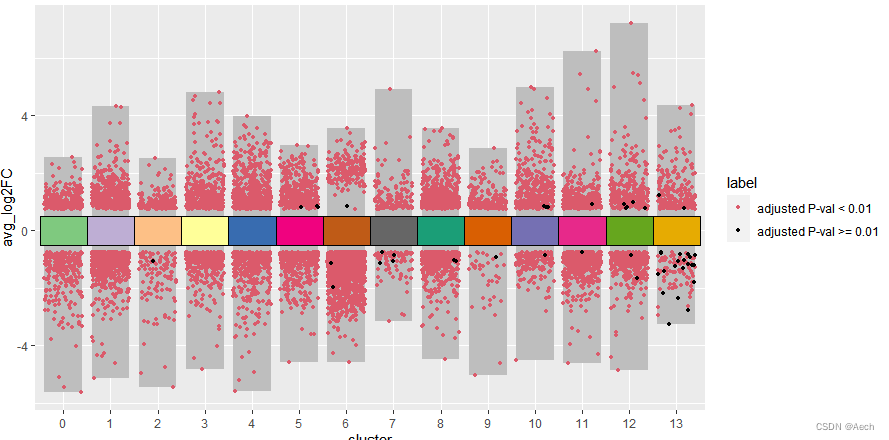
- 添加注释方块
ggplot(deg, aes(x = cluster, y = avg_log2FC ))+
geom_col(data = bardata, mapping = aes(x = cluster, y = tail),
fill = "grey", width = 0.8) +
geom_col(data = bardata, mapping = aes(x = cluster, y = top),
fill = "grey", width = 0.8) +
geom_jitter(aes(color = label), size = 1,
position = position_jitter(seed = 0328)) +
scale_color_manual(values = c("#db5a6b", "black")) +
geom_tile(aes(y = 0, fill = cluster), show.legend = F,
color = "black", width = 1) +
scale_fill_manual(values = col_vector)
- 给想要展示的基因和注释方块添加文字
- 看着有点挤, 点击zoom放大就好了
ggplot(deg, aes(x = cluster, y = avg_log2FC ))+
geom_col(data = bardata, mapping = aes(x = cluster, y = tail),
fill = "grey", width = 0.8) +
geom_col(data = bardata, mapping = aes(x = cluster, y = top),
fill = "grey", width = 0.8) +
geom_jitter(aes(color = label), size = 1,
position = position_jitter(seed = 0328)) +
scale_color_manual(values = c("#db5a6b", "black")) +
geom_tile(aes(y = 0, fill = cluster), show.legend = F,
color = "black", width = 1) +
scale_fill_manual(values = col_vector) +
geom_text(aes(y = 0, label = cluster)) +
geom_text_repel(data = deg %>% filter(gene %in% unique(tagedgene$gene)),
aes(label = gene), position = position_jitter(seed = 0328),
arrow = arrow(angle = 30, length = unit(0.05, "inches"),
ends = "last", type = "open"))

- 最后处理一下背景啥的
ggplot(deg, aes(x = cluster, y = avg_log2FC ))+
geom_col(data = bardata, mapping = aes(x = cluster, y = tail),
fill = "grey", width = 0.8) +
geom_col(data = bardata, mapping = aes(x = cluster, y = top),
fill = "grey", width = 0.8) +
geom_jitter(aes(color = label), size = 1,
position = position_jitter(seed = 0328)) +
scale_color_manual(values = c("#db5a6b", "black")) +
geom_tile(aes(y = 0, fill = cluster), show.legend = F,
color = "black", width = 1) +
scale_fill_manual(values = col_vector) +
geom_text(aes(y = 0, label = cluster)) +
geom_text_repel(data = deg %>% filter(gene %in% unique(tagedgene$gene)),
aes(label = gene), position = position_jitter(seed = 0328),
arrow = arrow(angle = 30, length = unit(0.05, "inches"),
ends = "last", type = "open")) +
theme_minimal() +
theme(axis.line.y = element_line(color = "black", linewidth = 1),
axis.line.x = element_blank(),
axis.text.x = element_blank(),
panel.grid = element_blank(),
legend.title = element_blank())
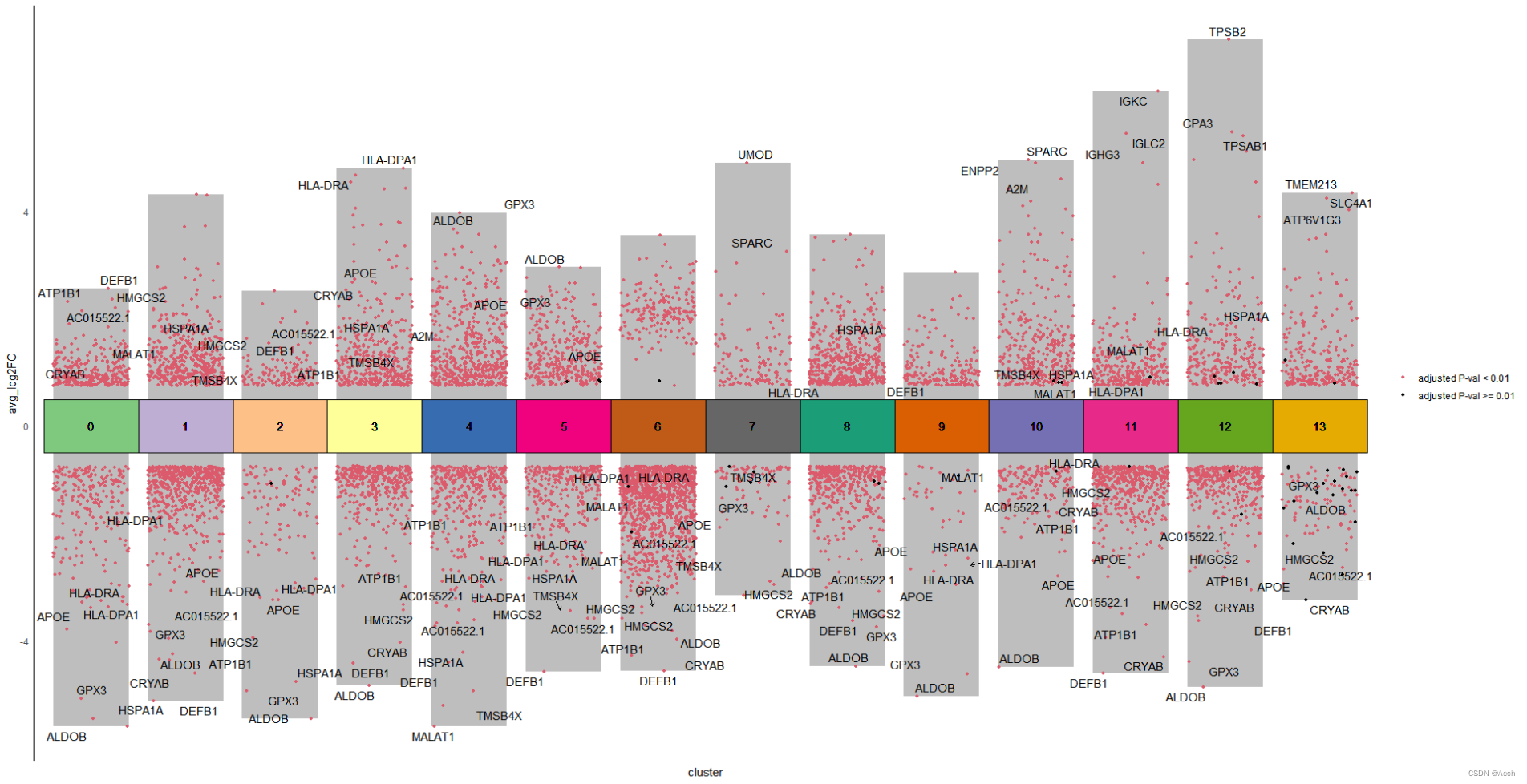
是不是很简单啊 😃
其实不只是单细胞, RNAseq等技术的差异基因也可以组合成类似的矩阵之后绘制相同的多组差异火山图. 理解这个图是柱状图和散点图的结合就可以灵活的绘制类似的图啦 😃
文章来源:https://blog.csdn.net/Aechh/article/details/135384734
本文来自互联网用户投稿,该文观点仅代表作者本人,不代表本站立场。本站仅提供信息存储空间服务,不拥有所有权,不承担相关法律责任。 如若内容造成侵权/违法违规/事实不符,请联系我的编程经验分享网邮箱:chenni525@qq.com进行投诉反馈,一经查实,立即删除!
本文来自互联网用户投稿,该文观点仅代表作者本人,不代表本站立场。本站仅提供信息存储空间服务,不拥有所有权,不承担相关法律责任。 如若内容造成侵权/违法违规/事实不符,请联系我的编程经验分享网邮箱:chenni525@qq.com进行投诉反馈,一经查实,立即删除!
最新文章
- Python教程
- 深入理解 MySQL 中的 HAVING 关键字和聚合函数
- Qt之QChar编码(1)
- MyBatis入门基础篇
- 用Python脚本实现FFmpeg批量转换
- Ubuntu开机自启动文件
- GoLang:gRPC协议
- 如何用 jigdo 下载 debian
- rtklib读取原始数据是一次读取了一个文件的全部数据
- 前端框架中的状态管理(State Management)
- 【数据结构】排序之归并排序与计数排序
- 快快销ShopMatrix 分销商城多端uniapp可编译5端 - 最早推荐的两个会员的团队业绩最小值
- 提供电商Api接口-100种接口,淘宝,1688,抖音商品详情数据安全,稳定,支持高并发
- acwing讲解篇之93. 递归实现组合型枚举
- 珠海翼型件自动化3D偏差检测分析抄数三维扫描数模全尺寸检测偏差