High-Throughput Algae Cell Detection
一、数据集
About Dataset
Microalgae, as an important natural resource, find widespread applications in various fields such as marine environments, biomedical research, clean energy, and food engineering. Monitoring the abundance and species composition of microalgae in water bodies can help identify potential environmental issues such as eutrophication, changes in water quality, and ecological balance. This is crucial for ecological assessment, establishing warning systems, and implementing the restoration of marine ecosystems. One major challenge in microalgae research lies in the rapid identification and quantification of diverse and variable microalgal samples. Currently, researchers mainly rely on microscopic examination of microalgal cells. However, this method often suffers from low throughput and sample damage, potentially overlooking the inherent heterogeneity among cells.
To address the above problem, microfluidic chip technology is a promising solution. Microfluidic chips are characterized by miniaturization, integration, high sensitivity and low cost, and have gradually become an ideal platform for real-time cell analysis and microbial monitoring. However, the existing cell detection models on microfluidic platforms have limitations, and it is difficult to achieve multi-classification of cells and accurate detection of cells with similar size but different states at the same time. This is especially evident in microalgae samples, due to the diversity of species, the size variation of cells within the same species, and the significant size difference between different species, which pose a unique challenge. Therefore, there is an urgent need to develop a label-free, high-throughput, multi-classification, multi-scale microalgae cell detection method to meet the practical needs of microalgae research. This development will help promote technological innovation in the microalgae field and provide more reliable tools for future ecological protection and resource management.
To address the challenges of high-throughput algae cell detection, we have organized the second International “Vision Meets Algae” (VisAlgae) Challenge and Workshop. VisAlgae 2023 will be held in conjunction with the IEEE Cybermatics Conference, focusing on the interdisciplinary application of algae research and computer vision technology. We conducted experiments on a high-throughput microfluidic platform, collecting dynamic video data of microalgal cells under different fields of view and imaging conditions. The experimental subjects include six types of microalgal cells: Platymonas, Chlorella, Dunaliella salina, Effrenium, Porphyridium, and Haematococcus. These cells exhibit significant differences in size, pose challenges with extremely small targets, and may experience motion blur and defocus under certain conditions. Our task is to address the practical challenges of high-throughput algae cell detection, develop targeted object detection algorithms to overcome issues such as detecting small targets, handling multiscale issues, managing motion blur, dealing with complex backgrounds, and maximize detection accuracy.
The training set contains 700 images and the testing set contains 300 images, with 6 classes. The annotations of training set are in YOLO format. Each image has a corresponding annotation file in .txt format.
Each line in the txt file means:Class, x_center, y_center, w, h
Classes names:
0: Platymonas
1: Chlorella
2: Dunaliella salina
3: Effrenium
4: Porphyridium
5: Haematococcus
二、yolov5训练结果
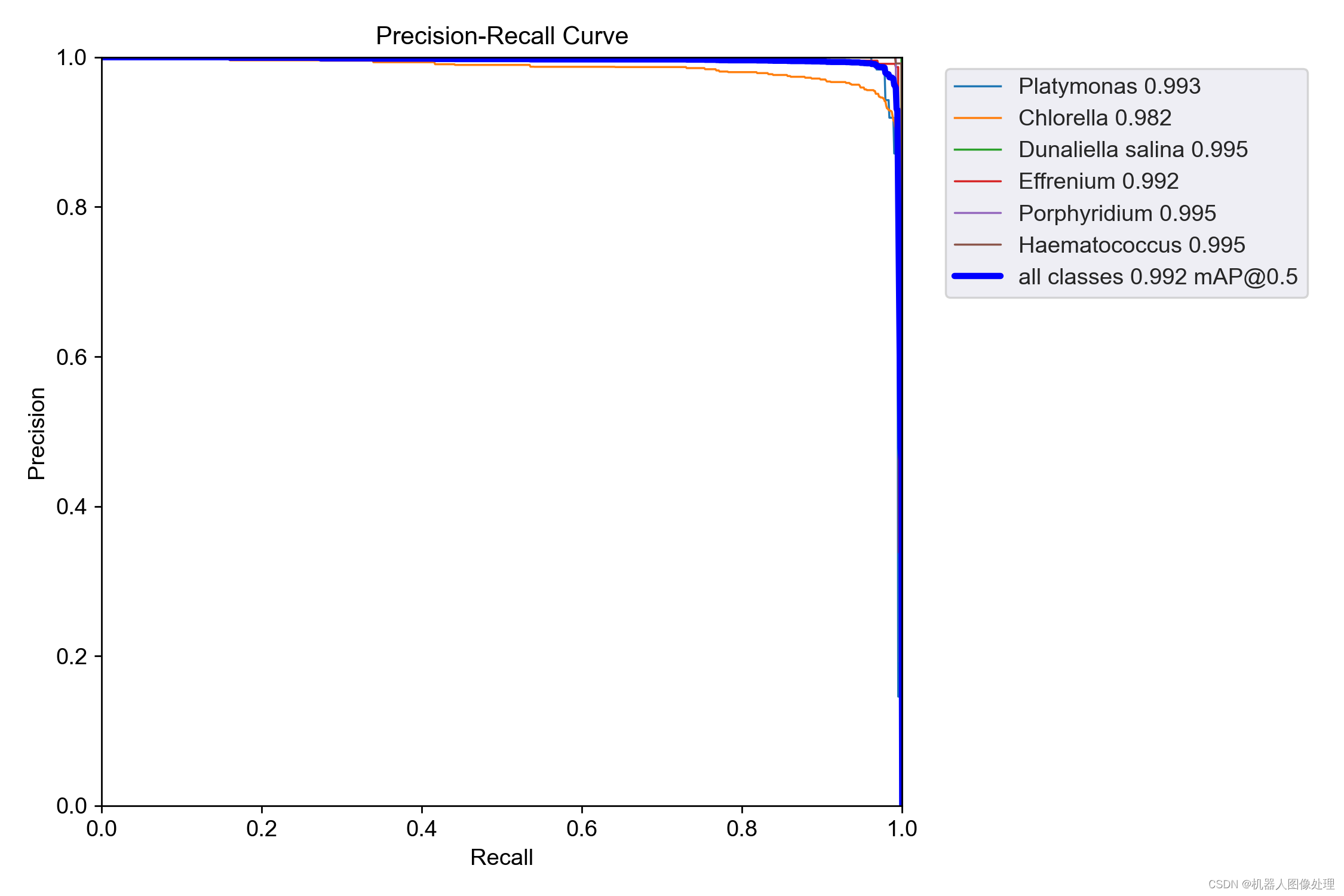
all 700 1758 0.987 0.985 0.992 0.811
Platymonas 700 184 0.99 0.967 0.993 0.833
Chlorella 700 722 0.951 0.969 0.982 0.641
Dunaliella salina 700 234 0.996 1 0.995 0.824
Effrenium 700 227 0.991 0.99 0.992 0.85
Porphyridium 700 272 0.996 0.993 0.995 0.809
Haematococcus 700 119 0.999 0.992 0.995 0.909
三、测试






本文来自互联网用户投稿,该文观点仅代表作者本人,不代表本站立场。本站仅提供信息存储空间服务,不拥有所有权,不承担相关法律责任。 如若内容造成侵权/违法违规/事实不符,请联系我的编程经验分享网邮箱:chenni525@qq.com进行投诉反馈,一经查实,立即删除!
- Python教程
- 深入理解 MySQL 中的 HAVING 关键字和聚合函数
- Qt之QChar编码(1)
- MyBatis入门基础篇
- 用Python脚本实现FFmpeg批量转换
- 基于Matlab的车道线检测技术研究与实现
- Poi实现excel报表导入,通过原始poi实现可应对各种格式的报表
- HTTPS协议把什么加密了?
- 联想发布天禧AI生态四端一体战略,聚焦智能体小程序开发
- 初识大数据,一文掌握大数据必备知识文集(3)
- Spring AOP
- 【MySQL】在 Centos7 环境下安装 MySQL
- pgAdmin 4的安装与使用
- 【C】const 关键字 xTaskCreate(const char * const pcName)
- 一致性哈希算法