工具系列:TensorFlow决策森林_(7)检查和调试决策森林模型
在本文中,您将学习如何直接检查和创建模型的结构。我们假设您已经熟悉了在初级和中级介绍的概念。
在本文中,您将:
-
训练一个随机森林模型并以编程方式访问其结构。
-
手动创建一个随机森林模型,并将其用作经典模型。
设置
# 安装 TensorFlow Decision Forests 库
!pip install tensorflow_decision_forests
# 安装 wurlitzer 库,用于显示训练日志
!pip install wurlitzer
Collecting tensorflow_decision_forests
Using cached tensorflow_decision_forests-1.1.0-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl (16.2 MB)
Requirement already satisfied: wheel in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorflow_decision_forests) (0.37.1)
Requirement already satisfied: numpy in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorflow_decision_forests) (1.24.0rc2)
Requirement already satisfied: absl-py in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorflow_decision_forests) (1.3.0)
Requirement already satisfied: six in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorflow_decision_forests) (1.16.0)
Collecting wurlitzer
Using cached wurlitzer-3.0.3-py3-none-any.whl (7.3 kB)
Requirement already satisfied: pandas in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorflow_decision_forests) (1.5.2)
Requirement already satisfied: tensorflow~=2.11.0 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorflow_decision_forests) (2.11.0)
Requirement already satisfied: setuptools in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorflow~=2.11.0->tensorflow_decision_forests) (65.6.3)
Requirement already satisfied: gast<=0.4.0,>=0.2.1 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorflow~=2.11.0->tensorflow_decision_forests) (0.4.0)
Requirement already satisfied: h5py>=2.9.0 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorflow~=2.11.0->tensorflow_decision_forests) (3.7.0)
Requirement already satisfied: libclang>=13.0.0 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorflow~=2.11.0->tensorflow_decision_forests) (14.0.6)
Requirement already satisfied: flatbuffers>=2.0 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorflow~=2.11.0->tensorflow_decision_forests) (22.12.6)
Requirement already satisfied: tensorboard<2.12,>=2.11 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorflow~=2.11.0->tensorflow_decision_forests) (2.11.0)
Requirement already satisfied: typing-extensions>=3.6.6 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorflow~=2.11.0->tensorflow_decision_forests) (4.4.0)
Requirement already satisfied: keras<2.12,>=2.11.0 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorflow~=2.11.0->tensorflow_decision_forests) (2.11.0)
Requirement already satisfied: wrapt>=1.11.0 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorflow~=2.11.0->tensorflow_decision_forests) (1.14.1)
Requirement already satisfied: astunparse>=1.6.0 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorflow~=2.11.0->tensorflow_decision_forests) (1.6.3)
Requirement already satisfied: tensorflow-estimator<2.12,>=2.11.0 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorflow~=2.11.0->tensorflow_decision_forests) (2.11.0)
Requirement already satisfied: protobuf<3.20,>=3.9.2 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorflow~=2.11.0->tensorflow_decision_forests) (3.19.6)
Requirement already satisfied: opt-einsum>=2.3.2 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorflow~=2.11.0->tensorflow_decision_forests) (3.3.0)
Requirement already satisfied: grpcio<2.0,>=1.24.3 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorflow~=2.11.0->tensorflow_decision_forests) (1.51.1)
Requirement already satisfied: packaging in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorflow~=2.11.0->tensorflow_decision_forests) (22.0)
Requirement already satisfied: termcolor>=1.1.0 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorflow~=2.11.0->tensorflow_decision_forests) (2.1.1)
Requirement already satisfied: tensorflow-io-gcs-filesystem>=0.23.1 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorflow~=2.11.0->tensorflow_decision_forests) (0.28.0)
Requirement already satisfied: google-pasta>=0.1.1 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorflow~=2.11.0->tensorflow_decision_forests) (0.2.0)
Requirement already satisfied: python-dateutil>=2.8.1 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from pandas->tensorflow_decision_forests) (2.8.2)
Requirement already satisfied: pytz>=2020.1 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from pandas->tensorflow_decision_forests) (2022.6)
Requirement already satisfied: werkzeug>=1.0.1 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorboard<2.12,>=2.11->tensorflow~=2.11.0->tensorflow_decision_forests) (2.2.2)
Requirement already satisfied: markdown>=2.6.8 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorboard<2.12,>=2.11->tensorflow~=2.11.0->tensorflow_decision_forests) (3.4.1)
Requirement already satisfied: tensorboard-plugin-wit>=1.6.0 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorboard<2.12,>=2.11->tensorflow~=2.11.0->tensorflow_decision_forests) (1.8.1)
Requirement already satisfied: tensorboard-data-server<0.7.0,>=0.6.0 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorboard<2.12,>=2.11->tensorflow~=2.11.0->tensorflow_decision_forests) (0.6.1)
Requirement already satisfied: requests<3,>=2.21.0 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorboard<2.12,>=2.11->tensorflow~=2.11.0->tensorflow_decision_forests) (2.28.1)
Requirement already satisfied: google-auth-oauthlib<0.5,>=0.4.1 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorboard<2.12,>=2.11->tensorflow~=2.11.0->tensorflow_decision_forests) (0.4.6)
Requirement already satisfied: google-auth<3,>=1.6.3 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from tensorboard<2.12,>=2.11->tensorflow~=2.11.0->tensorflow_decision_forests) (2.15.0)
Requirement already satisfied: cachetools<6.0,>=2.0.0 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from google-auth<3,>=1.6.3->tensorboard<2.12,>=2.11->tensorflow~=2.11.0->tensorflow_decision_forests) (5.2.0)
Requirement already satisfied: rsa<5,>=3.1.4 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from google-auth<3,>=1.6.3->tensorboard<2.12,>=2.11->tensorflow~=2.11.0->tensorflow_decision_forests) (4.9)
Requirement already satisfied: pyasn1-modules>=0.2.1 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from google-auth<3,>=1.6.3->tensorboard<2.12,>=2.11->tensorflow~=2.11.0->tensorflow_decision_forests) (0.3.0rc1)
Requirement already satisfied: requests-oauthlib>=0.7.0 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from google-auth-oauthlib<0.5,>=0.4.1->tensorboard<2.12,>=2.11->tensorflow~=2.11.0->tensorflow_decision_forests) (1.3.1)
Requirement already satisfied: importlib-metadata>=4.4 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from markdown>=2.6.8->tensorboard<2.12,>=2.11->tensorflow~=2.11.0->tensorflow_decision_forests) (5.1.0)
Requirement already satisfied: charset-normalizer<3,>=2 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from requests<3,>=2.21.0->tensorboard<2.12,>=2.11->tensorflow~=2.11.0->tensorflow_decision_forests) (2.1.1)
Requirement already satisfied: idna<4,>=2.5 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from requests<3,>=2.21.0->tensorboard<2.12,>=2.11->tensorflow~=2.11.0->tensorflow_decision_forests) (3.4)
Requirement already satisfied: certifi>=2017.4.17 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from requests<3,>=2.21.0->tensorboard<2.12,>=2.11->tensorflow~=2.11.0->tensorflow_decision_forests) (2022.12.7)
Requirement already satisfied: urllib3<1.27,>=1.21.1 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from requests<3,>=2.21.0->tensorboard<2.12,>=2.11->tensorflow~=2.11.0->tensorflow_decision_forests) (1.26.13)
Requirement already satisfied: MarkupSafe>=2.1.1 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from werkzeug>=1.0.1->tensorboard<2.12,>=2.11->tensorflow~=2.11.0->tensorflow_decision_forests) (2.1.1)
Requirement already satisfied: zipp>=0.5 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from importlib-metadata>=4.4->markdown>=2.6.8->tensorboard<2.12,>=2.11->tensorflow~=2.11.0->tensorflow_decision_forests) (3.11.0)
Requirement already satisfied: pyasn1<0.6.0,>=0.4.6 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from pyasn1-modules>=0.2.1->google-auth<3,>=1.6.3->tensorboard<2.12,>=2.11->tensorflow~=2.11.0->tensorflow_decision_forests) (0.5.0rc2)
Requirement already satisfied: oauthlib>=3.0.0 in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (from requests-oauthlib>=0.7.0->google-auth-oauthlib<0.5,>=0.4.1->tensorboard<2.12,>=2.11->tensorflow~=2.11.0->tensorflow_decision_forests) (3.2.2)
Installing collected packages: wurlitzer, tensorflow_decision_forests
Successfully installed tensorflow_decision_forests-1.1.0 wurlitzer-3.0.3
Requirement already satisfied: wurlitzer in /tmpfs/src/tf_docs_env/lib/python3.9/site-packages (3.0.3)
# 导入tensorflow_decision_forests库
import tensorflow_decision_forests as tfdf
# 导入os、numpy、pandas、tensorflow、matplotlib.pyplot、math、collections库
import os
import numpy as np
import pandas as pd
import tensorflow as tf
import matplotlib.pyplot as plt
import math
import collections
2022-12-14 12:24:51.050867: W tensorflow/compiler/xla/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libnvinfer.so.7'; dlerror: libnvinfer.so.7: cannot open shared object file: No such file or directory
2022-12-14 12:24:51.050964: W tensorflow/compiler/xla/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libnvinfer_plugin.so.7'; dlerror: libnvinfer_plugin.so.7: cannot open shared object file: No such file or directory
2022-12-14 12:24:51.050973: W tensorflow/compiler/tf2tensorrt/utils/py_utils.cc:38] TF-TRT Warning: Cannot dlopen some TensorRT libraries. If you would like to use Nvidia GPU with TensorRT, please make sure the missing libraries mentioned above are installed properly.
隐藏的代码单元格限制了在colab中的输出高度。
# 导入所需的模块
from IPython.core.magic import register_line_magic
from IPython.display import Javascript
from IPython.display import display as ipy_display
# 定义一个魔术命令,用于设置单元格的最大高度
@register_line_magic
def set_cell_height(size):
# 调用Javascript代码,设置单元格的最大高度
ipy_display(
Javascript("google.colab.output.setIframeHeight(0, true, {maxHeight: " +
str(size) + "})"))
训练一个简单的随机森林
我们像在初学者colab中一样训练一个随机森林。
# 下载数据集
!wget -q https://storage.googleapis.com/download.tensorflow.org/data/palmer_penguins/penguins.csv -O /tmp/penguins.csv
# 将数据集加载到Pandas Dataframe中
dataset_df = pd.read_csv("/tmp/penguins.csv")
# 显示前三个示例
print(dataset_df.head(3))
# 将Pandas Dataframe转换为tf数据集
dataset_tf = tfdf.keras.pd_dataframe_to_tf_dataset(dataset_df, label="species")
# 训练随机森林模型
model = tfdf.keras.RandomForestModel(compute_oob_variable_importances=True)
model.fit(x=dataset_tf)
species island bill_length_mm bill_depth_mm flipper_length_mm \
0 Adelie Torgersen 39.1 18.7 181.0
1 Adelie Torgersen 39.5 17.4 186.0
2 Adelie Torgersen 40.3 18.0 195.0
body_mass_g sex year
0 3750.0 male 2007
1 3800.0 female 2007
2 3250.0 female 2007
Warning: The `num_threads` constructor argument is not set and the number of CPU is os.cpu_count()=32 > 32. Setting num_threads to 32. Set num_threads manually to use more than 32 cpus.
WARNING:absl:The `num_threads` constructor argument is not set and the number of CPU is os.cpu_count()=32 > 32. Setting num_threads to 32. Set num_threads manually to use more than 32 cpus.
Use /tmpfs/tmp/tmpvr7urazn as temporary training directory
Reading training dataset...
WARNING:tensorflow:From /tmpfs/src/tf_docs_env/lib/python3.9/site-packages/tensorflow/python/autograph/pyct/static_analysis/liveness.py:83: Analyzer.lamba_check (from tensorflow.python.autograph.pyct.static_analysis.liveness) is deprecated and will be removed after 2023-09-23.
Instructions for updating:
Lambda fuctions will be no more assumed to be used in the statement where they are used, or at least in the same block. https://github.com/tensorflow/tensorflow/issues/56089
WARNING:tensorflow:From /tmpfs/src/tf_docs_env/lib/python3.9/site-packages/tensorflow/python/autograph/pyct/static_analysis/liveness.py:83: Analyzer.lamba_check (from tensorflow.python.autograph.pyct.static_analysis.liveness) is deprecated and will be removed after 2023-09-23.
Instructions for updating:
Lambda fuctions will be no more assumed to be used in the statement where they are used, or at least in the same block. https://github.com/tensorflow/tensorflow/issues/56089
Training dataset read in 0:00:02.961832. Found 344 examples.
Training model...
Model trained in 0:00:00.093680
Compiling model...
[INFO 2022-12-14T12:24:58.955519768+00:00 kernel.cc:1175] Loading model from path /tmpfs/tmp/tmpvr7urazn/model/ with prefix fb8057db01324481
[INFO 2022-12-14T12:24:58.971817533+00:00 abstract_model.cc:1306] Engine "RandomForestGeneric" built
[INFO 2022-12-14T12:24:58.97187255+00:00 kernel.cc:1021] Use fast generic engine
WARNING:tensorflow:AutoGraph could not transform <function simple_ml_inference_op_with_handle at 0x7f9b54f644c0> and will run it as-is.
Please report this to the TensorFlow team. When filing the bug, set the verbosity to 10 (on Linux, `export AUTOGRAPH_VERBOSITY=10`) and attach the full output.
Cause: could not get source code
To silence this warning, decorate the function with @tf.autograph.experimental.do_not_convert
WARNING:tensorflow:AutoGraph could not transform <function simple_ml_inference_op_with_handle at 0x7f9b54f644c0> and will run it as-is.
Please report this to the TensorFlow team. When filing the bug, set the verbosity to 10 (on Linux, `export AUTOGRAPH_VERBOSITY=10`) and attach the full output.
Cause: could not get source code
To silence this warning, decorate the function with @tf.autograph.experimental.do_not_convert
WARNING: AutoGraph could not transform <function simple_ml_inference_op_with_handle at 0x7f9b54f644c0> and will run it as-is.
Please report this to the TensorFlow team. When filing the bug, set the verbosity to 10 (on Linux, `export AUTOGRAPH_VERBOSITY=10`) and attach the full output.
Cause: could not get source code
To silence this warning, decorate the function with @tf.autograph.experimental.do_not_convert
Model compiled.
<keras.callbacks.History at 0x7f9b5394c6d0>
请注意模型构造函数中的compute_oob_variable_importances=True超参数。此选项在训练过程中计算袋外(OOB)变量重要性。这是随机森林模型的一种流行的排列变量重要性。
计算OOB变量重要性不会影响最终模型,但会减慢大型数据集的训练速度。
请检查模型摘要:
# 打印模型的概述信息
model.summary()
<IPython.core.display.Javascript object>
Model: "random_forest_model"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
=================================================================
Total params: 1
Trainable params: 0
Non-trainable params: 1
_________________________________________________________________
Type: "RANDOM_FOREST"
Task: CLASSIFICATION
Label: "__LABEL"
Input Features (7):
bill_depth_mm
bill_length_mm
body_mass_g
flipper_length_mm
island
sex
year
No weights
Variable Importance: MEAN_DECREASE_IN_ACCURACY:
1. "bill_length_mm" 0.151163 ################
2. "island" 0.008721 #
3. "bill_depth_mm" 0.000000
4. "body_mass_g" 0.000000
5. "sex" 0.000000
6. "year" 0.000000
7. "flipper_length_mm" -0.002907
Variable Importance: MEAN_DECREASE_IN_AP_1_VS_OTHERS:
1. "bill_length_mm" 0.083305 ################
2. "island" 0.007664 #
3. "flipper_length_mm" 0.003400
4. "bill_depth_mm" 0.002741
5. "body_mass_g" 0.000722
6. "sex" 0.000644
7. "year" 0.000000
Variable Importance: MEAN_DECREASE_IN_AP_2_VS_OTHERS:
1. "bill_length_mm" 0.508510 ################
2. "island" 0.023487
3. "bill_depth_mm" 0.007744
4. "flipper_length_mm" 0.006008
5. "body_mass_g" 0.003017
6. "sex" 0.001537
7. "year" -0.000245
Variable Importance: MEAN_DECREASE_IN_AP_3_VS_OTHERS:
1. "island" 0.002192 ################
2. "bill_length_mm" 0.001572 ############
3. "bill_depth_mm" 0.000497 #######
4. "sex" 0.000000 ####
5. "year" 0.000000 ####
6. "body_mass_g" -0.000053 ####
7. "flipper_length_mm" -0.000890
Variable Importance: MEAN_DECREASE_IN_AUC_1_VS_OTHERS:
1. "bill_length_mm" 0.071306 ################
2. "island" 0.007299 #
3. "flipper_length_mm" 0.004506 #
4. "bill_depth_mm" 0.002124
5. "body_mass_g" 0.000548
6. "sex" 0.000480
7. "year" 0.000000
Variable Importance: MEAN_DECREASE_IN_AUC_2_VS_OTHERS:
1. "bill_length_mm" 0.108642 ################
2. "island" 0.014493 ##
3. "bill_depth_mm" 0.007406 #
4. "flipper_length_mm" 0.005195
5. "body_mass_g" 0.001012
6. "sex" 0.000480
7. "year" -0.000053
Variable Importance: MEAN_DECREASE_IN_AUC_3_VS_OTHERS:
1. "island" 0.002126 ################
2. "bill_length_mm" 0.001393 ###########
3. "bill_depth_mm" 0.000293 #####
4. "sex" 0.000000 ###
5. "year" 0.000000 ###
6. "body_mass_g" -0.000037 ###
7. "flipper_length_mm" -0.000550
Variable Importance: MEAN_DECREASE_IN_PRAUC_1_VS_OTHERS:
1. "bill_length_mm" 0.083122 ################
2. "island" 0.010887 ##
3. "flipper_length_mm" 0.003425
4. "bill_depth_mm" 0.002731
5. "body_mass_g" 0.000719
6. "sex" 0.000641
7. "year" 0.000000
Variable Importance: MEAN_DECREASE_IN_PRAUC_2_VS_OTHERS:
1. "bill_length_mm" 0.497611 ################
2. "island" 0.024045
3. "bill_depth_mm" 0.007734
4. "flipper_length_mm" 0.006017
5. "body_mass_g" 0.003000
6. "sex" 0.001528
7. "year" -0.000243
Variable Importance: MEAN_DECREASE_IN_PRAUC_3_VS_OTHERS:
1. "island" 0.002187 ################
2. "bill_length_mm" 0.001568 ############
3. "bill_depth_mm" 0.000495 #######
4. "sex" 0.000000 ####
5. "year" 0.000000 ####
6. "body_mass_g" -0.000053 ####
7. "flipper_length_mm" -0.000886
Variable Importance: MEAN_MIN_DEPTH:
1. "__LABEL" 3.479602 ################
2. "year" 3.463891 ###############
3. "sex" 3.430498 ###############
4. "body_mass_g" 2.898112 ###########
5. "island" 2.388925 ########
6. "bill_depth_mm" 2.336100 #######
7. "bill_length_mm" 1.282960
8. "flipper_length_mm" 1.270079
Variable Importance: NUM_AS_ROOT:
1. "flipper_length_mm" 157.000000 ################
2. "bill_length_mm" 76.000000 #######
3. "bill_depth_mm" 52.000000 #####
4. "island" 12.000000
5. "body_mass_g" 3.000000
Variable Importance: NUM_NODES:
1. "bill_length_mm" 778.000000 ################
2. "bill_depth_mm" 463.000000 #########
3. "flipper_length_mm" 414.000000 ########
4. "island" 342.000000 ######
5. "body_mass_g" 338.000000 ######
6. "sex" 36.000000
7. "year" 19.000000
Variable Importance: SUM_SCORE:
1. "bill_length_mm" 36515.793787 ################
2. "flipper_length_mm" 35120.434174 ###############
3. "island" 14669.408395 ######
4. "bill_depth_mm" 14515.446617 ######
5. "body_mass_g" 3485.330881 #
6. "sex" 354.201073
7. "year" 49.737758
Winner takes all: true
Out-of-bag evaluation: accuracy:0.976744 logloss:0.0678223
Number of trees: 300
Total number of nodes: 5080
Number of nodes by tree:
Count: 300 Average: 16.9333 StdDev: 3.10197
Min: 11 Max: 31 Ignored: 0
----------------------------------------------
[ 11, 12) 6 2.00% 2.00% #
[ 12, 13) 0 0.00% 2.00%
[ 13, 14) 46 15.33% 17.33% #####
[ 14, 15) 0 0.00% 17.33%
[ 15, 16) 70 23.33% 40.67% ########
[ 16, 17) 0 0.00% 40.67%
[ 17, 18) 84 28.00% 68.67% ##########
[ 18, 19) 0 0.00% 68.67%
[ 19, 20) 46 15.33% 84.00% #####
[ 20, 21) 0 0.00% 84.00%
[ 21, 22) 30 10.00% 94.00% ####
[ 22, 23) 0 0.00% 94.00%
[ 23, 24) 13 4.33% 98.33% ##
[ 24, 25) 0 0.00% 98.33%
[ 25, 26) 2 0.67% 99.00%
[ 26, 27) 0 0.00% 99.00%
[ 27, 28) 2 0.67% 99.67%
[ 28, 29) 0 0.00% 99.67%
[ 29, 30) 0 0.00% 99.67%
[ 30, 31] 1 0.33% 100.00%
Depth by leafs:
Count: 2690 Average: 3.53271 StdDev: 1.06789
Min: 2 Max: 7 Ignored: 0
----------------------------------------------
[ 2, 3) 545 20.26% 20.26% ######
[ 3, 4) 747 27.77% 48.03% ########
[ 4, 5) 888 33.01% 81.04% ##########
[ 5, 6) 444 16.51% 97.55% #####
[ 6, 7) 62 2.30% 99.85% #
[ 7, 7] 4 0.15% 100.00%
Number of training obs by leaf:
Count: 2690 Average: 38.3643 StdDev: 44.8651
Min: 5 Max: 155 Ignored: 0
----------------------------------------------
[ 5, 12) 1474 54.80% 54.80% ##########
[ 12, 20) 124 4.61% 59.41% #
[ 20, 27) 48 1.78% 61.19%
[ 27, 35) 74 2.75% 63.94% #
[ 35, 42) 58 2.16% 66.10%
[ 42, 50) 85 3.16% 69.26% #
[ 50, 57) 96 3.57% 72.83% #
[ 57, 65) 87 3.23% 76.06% #
[ 65, 72) 49 1.82% 77.88%
[ 72, 80) 23 0.86% 78.74%
[ 80, 88) 30 1.12% 79.85%
[ 88, 95) 23 0.86% 80.71%
[ 95, 103) 42 1.56% 82.27%
[ 103, 110) 62 2.30% 84.57%
[ 110, 118) 115 4.28% 88.85% #
[ 118, 125) 115 4.28% 93.12% #
[ 125, 133) 98 3.64% 96.77% #
[ 133, 140) 49 1.82% 98.59%
[ 140, 148) 31 1.15% 99.74%
[ 148, 155] 7 0.26% 100.00%
Attribute in nodes:
778 : bill_length_mm [NUMERICAL]
463 : bill_depth_mm [NUMERICAL]
414 : flipper_length_mm [NUMERICAL]
342 : island [CATEGORICAL]
338 : body_mass_g [NUMERICAL]
36 : sex [CATEGORICAL]
19 : year [NUMERICAL]
Attribute in nodes with depth <= 0:
157 : flipper_length_mm [NUMERICAL]
76 : bill_length_mm [NUMERICAL]
52 : bill_depth_mm [NUMERICAL]
12 : island [CATEGORICAL]
3 : body_mass_g [NUMERICAL]
Attribute in nodes with depth <= 1:
250 : bill_length_mm [NUMERICAL]
244 : flipper_length_mm [NUMERICAL]
183 : bill_depth_mm [NUMERICAL]
170 : island [CATEGORICAL]
53 : body_mass_g [NUMERICAL]
Attribute in nodes with depth <= 2:
462 : bill_length_mm [NUMERICAL]
320 : flipper_length_mm [NUMERICAL]
310 : bill_depth_mm [NUMERICAL]
287 : island [CATEGORICAL]
162 : body_mass_g [NUMERICAL]
9 : sex [CATEGORICAL]
5 : year [NUMERICAL]
Attribute in nodes with depth <= 3:
669 : bill_length_mm [NUMERICAL]
410 : bill_depth_mm [NUMERICAL]
383 : flipper_length_mm [NUMERICAL]
328 : island [CATEGORICAL]
286 : body_mass_g [NUMERICAL]
32 : sex [CATEGORICAL]
10 : year [NUMERICAL]
Attribute in nodes with depth <= 5:
778 : bill_length_mm [NUMERICAL]
462 : bill_depth_mm [NUMERICAL]
413 : flipper_length_mm [NUMERICAL]
342 : island [CATEGORICAL]
338 : body_mass_g [NUMERICAL]
36 : sex [CATEGORICAL]
19 : year [NUMERICAL]
Condition type in nodes:
2012 : HigherCondition
378 : ContainsBitmapCondition
Condition type in nodes with depth <= 0:
288 : HigherCondition
12 : ContainsBitmapCondition
Condition type in nodes with depth <= 1:
730 : HigherCondition
170 : ContainsBitmapCondition
Condition type in nodes with depth <= 2:
1259 : HigherCondition
296 : ContainsBitmapCondition
Condition type in nodes with depth <= 3:
1758 : HigherCondition
360 : ContainsBitmapCondition
Condition type in nodes with depth <= 5:
2010 : HigherCondition
378 : ContainsBitmapCondition
Node format: NOT_SET
Training OOB:
trees: 1, Out-of-bag evaluation: accuracy:0.964286 logloss:1.28727
trees: 13, Out-of-bag evaluation: accuracy:0.959064 logloss:0.4869
trees: 31, Out-of-bag evaluation: accuracy:0.95614 logloss:0.284603
trees: 54, Out-of-bag evaluation: accuracy:0.973837 logloss:0.175283
trees: 73, Out-of-bag evaluation: accuracy:0.97093 logloss:0.175816
trees: 85, Out-of-bag evaluation: accuracy:0.973837 logloss:0.171781
trees: 96, Out-of-bag evaluation: accuracy:0.97093 logloss:0.077417
trees: 116, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0761788
trees: 127, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0745239
trees: 137, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0753508
trees: 150, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0741464
trees: 160, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0749481
trees: 170, Out-of-bag evaluation: accuracy:0.979651 logloss:0.0719624
trees: 190, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0711787
trees: 203, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0701121
trees: 213, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0682979
trees: 224, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0689686
trees: 248, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0674086
trees: 260, Out-of-bag evaluation: accuracy:0.976744 logloss:0.068218
trees: 270, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0680733
trees: 280, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0685965
trees: 290, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0683421
trees: 300, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0678223
注意,变量重要性有多个名称为MEAN_DECREASE_IN_*。
绘制模型
接下来,绘制模型。
随机森林是一个庞大的模型(该模型有300棵树和约5k个节点;请参见上面的摘要)。因此,只绘制第一棵树,并将节点限制在深度3。
# 使用model_plotter模块中的plot_model_in_colab函数来绘制模型
# 参数model表示要绘制的模型
# 参数tree_idx表示要绘制的树的索引,这里设置为0表示绘制第一棵树
# 参数max_depth表示要绘制的树的最大深度,这里设置为3表示绘制到第三层
tfdf.model_plotter.plot_model_in_colab(model, tree_idx=0, max_depth=3)
/**
- Plotting of decision trees generated by TF-DF.
- A tree is a recursive structure of node objects.
- A node contains one or more of the following components:
-
- A value: Representing the output of the node. If the node is not a leaf,
-
the value is only present for analysis i.e. it is not used for -
predictions. -
- A condition : For non-leaf nodes, the condition (also known as split)
-
defines a binary test to branch to the positive or negative child. -
- An explanation: Generally a plot showing the relation between the label
-
and the condition to give insights about the effect of the condition. -
- Two children : For non-leaf nodes, the children nodes. The first
-
children (i.e. "node.children[0]") is the negative children (drawn in -
red). The second children is the positive one (drawn in green).
*/
/**
- Plots a single decision tree into a DOM element.
- @param {!options} options Dictionary of configurations.
- @param {!tree} raw_tree Recursive tree structure.
- @param {string} canvas_id Id of the output dom element.
*/
function display_tree(options, raw_tree, canvas_id) {
console.log(options);
// Determine the node placement.
const tree_struct = d3.tree().nodeSize(
[options.node_y_offset, options.node_x_offset])(d3.hierarchy(raw_tree));
// Boundaries of the node placement.
let x_min = Infinity;
let x_max = -x_min;
let y_min = Infinity;
let y_max = -x_min;
tree_struct.each(d => {
if (d.x > x_max) x_max = d.x;
if (d.x < x_min) x_min = d.x;
if (d.y > y_max) y_max = d.y;
if (d.y < y_min) y_min = d.y;
});
// Size of the plot.
const width = y_max - y_min + options.node_x_size + options.margin * 2;
const height = x_max - x_min + options.node_y_size + options.margin * 2 +
options.node_y_offset - options.node_y_size;
const plot = d3.select(canvas_id);
// Tool tip
options.tooltip = plot.append(‘div’)
.attr(‘width’, 100)
.attr(‘height’, 100)
.style(‘padding’, ‘4px’)
.style(‘background’, ‘#fff’)
.style(‘box-shadow’, ‘4px 4px 0px rgba(0,0,0,0.1)’)
.style(‘border’, ‘1px solid black’)
.style(‘font-family’, ‘sans-serif’)
.style(‘font-size’, options.font_size)
.style(‘position’, ‘absolute’)
.style(‘z-index’, ‘10’)
.attr(‘pointer-events’, ‘none’)
.style(‘display’, ‘none’);
// Create canvas
const svg = plot.append(‘svg’).attr(‘width’, width).attr(‘height’, height);
const graph =
svg.style(‘overflow’, ‘visible’)
.append(‘g’)
.attr(‘font-family’, ‘sans-serif’)
.attr(‘font-size’, options.font_size)
.attr(
‘transform’,
() => translate(${options.margin},${ - x_min + options.node_y_offset / 2 + options.margin}));
// Plot bounding box.
if (options.show_plot_bounding_box) {
svg.append(‘rect’)
.attr(‘width’, width)
.attr(‘height’, height)
.attr(‘fill’, ‘none’)
.attr(‘stroke-width’, 1.0)
.attr(‘stroke’, ‘black’);
}
// Draw the edges.
display_edges(options, graph, tree_struct);
// Draw the nodes.
display_nodes(options, graph, tree_struct);
}
/**
- Draw the nodes of the tree.
- @param {!options} options Dictionary of configurations.
- @param {!graph} graph D3 search handle containing the graph.
- @param {!tree_struct} tree_struct Structure of the tree (node placement,
-
data, etc.).
*/
function display_nodes(options, graph, tree_struct) {
const nodes = graph.append(‘g’)
.selectAll(‘g’)
.data(tree_struct.descendants())
.join(‘g’)
.attr(‘transform’, d => translate(${d.y},${d.x}));
nodes.append(‘rect’)
.attr(‘x’, 0.5)
.attr(‘y’, 0.5)
.attr(‘width’, options.node_x_size)
.attr(‘height’, options.node_y_size)
.attr(‘stroke’, ‘lightgrey’)
.attr(‘stroke-width’, 1)
.attr(‘fill’, ‘white’)
.attr(‘y’, -options.node_y_size / 2);
// Brackets on the right of condition nodes without children.
non_leaf_node_without_children =
nodes.filter(node => node.data.condition != null && node.children == null)
.append(‘g’)
.attr(‘transform’, translate(${options.node_x_size},0));
non_leaf_node_without_children.append(‘path’)
.attr(‘d’, ‘M0,0 C 10,0 0,10 10,10’)
.attr(‘fill’, ‘none’)
.attr(‘stroke-width’, 1.0)
.attr(‘stroke’, ‘#F00’);
non_leaf_node_without_children.append(‘path’)
.attr(‘d’, ‘M0,0 C 10,0 0,-10 10,-10’)
.attr(‘fill’, ‘none’)
.attr(‘stroke-width’, 1.0)
.attr(‘stroke’, ‘#0F0’);
const node_content = nodes.append(‘g’).attr(
‘transform’,
translate(0,${options.node_padding - options.node_y_size / 2}));
node_content.append(node => create_node_element(options, node));
}
/**
- Creates the D3 content for a single node.
- @param {!options} options Dictionary of configurations.
- @param {!node} node Node to draw.
- @return {!d3} D3 content.
*/
function create_node_element(options, node) {
// Output accumulator.
let output = {
// Content to draw.
content: d3.create(‘svg:g’),
// Vertical offset to the next element to draw.
vertical_offset: 0
};
// Conditions.
if (node.data.condition != null) {
display_condition(options, node.data.condition, output);
}
// Values.
if (node.data.value != null) {
display_value(options, node.data.value, output);
}
// Explanations.
if (node.data.explanation != null) {
display_explanation(options, node.data.explanation, output);
}
return output.content.node();
}
/**
- Adds a single line of text inside of a node.
- @param {!options} options Dictionary of configurations.
- @param {string} text Text to display.
- @param {!output} output Output display accumulator.
*/
function display_node_text(options, text, output) {
output.content.append(‘text’)
.attr(‘x’, options.node_padding)
.attr(‘y’, output.vertical_offset)
.attr(‘alignment-baseline’, ‘hanging’)
.text(text);
output.vertical_offset += 10;
}
/**
- Adds a single line of text inside of a node with a tooltip.
- @param {!options} options Dictionary of configurations.
- @param {string} text Text to display.
- @param {string} tooltip Text in the Tooltip.
- @param {!output} output Output display accumulator.
*/
function display_node_text_with_tooltip(options, text, tooltip, output) {
const item = output.content.append(‘text’)
.attr(‘x’, options.node_padding)
.attr(‘alignment-baseline’, ‘hanging’)
.text(text);
add_tooltip(options, item, () => tooltip);
output.vertical_offset += 10;
}
/**
- Adds a tooltip to a dom element.
- @param {!options} options Dictionary of configurations.
- @param {!dom} target Dom element to equip with a tooltip.
- @param {!func} get_content Generates the html content of the tooltip.
*/
function add_tooltip(options, target, get_content) {
function show(d) {
options.tooltip.style(‘display’, ‘block’);
options.tooltip.html(get_content());
}
function hide(d) {
options.tooltip.style(‘display’, ‘none’);
}
function move(d) {
options.tooltip.style(‘display’, ‘block’);
options.tooltip.style(‘left’, (d.pageX + 5) + ‘px’);
options.tooltip.style(‘top’, d.pageY + ‘px’);
}
target.on(‘mouseover’, show);
target.on(‘mouseout’, hide);
target.on(‘mousemove’, move);
}
/**
- Adds a condition inside of a node.
- @param {!options} options Dictionary of configurations.
- @param {!condition} condition Condition to display.
- @param {!output} output Output display accumulator.
*/
function display_condition(options, condition, output) {
threshold_format = d3.format(‘r’);
if (condition.type === ‘IS_MISSING’) {
display_node_text(options, ${condition.attribute} is missing, output);
return;
}
if (condition.type === ‘IS_TRUE’) {
display_node_text(options, ${condition.attribute} is true, output);
return;
}
if (condition.type === ‘NUMERICAL_IS_HIGHER_THAN’) {
format = d3.format(‘r’);
display_node_text(
options,
${condition.attribute} >= ${threshold_format(condition.threshold)},
output);
return;
}
if (condition.type === ‘CATEGORICAL_IS_IN’) {
display_node_text_with_tooltip(
options, ${condition.attribute} in [...],
${condition.attribute} in [${condition.mask}], output);
return;
}
if (condition.type === ‘CATEGORICAL_SET_CONTAINS’) {
display_node_text_with_tooltip(
options, ${condition.attribute} intersect [...],
${condition.attribute} intersect [${condition.mask}], output);
return;
}
if (condition.type === ‘NUMERICAL_SPARSE_OBLIQUE’) {
display_node_text_with_tooltip(
options, Sparse oblique split...,
[${condition.attributes}]*[${condition.weights}]>=${ threshold_format(condition.threshold)},
output);
return;
}
display_node_text(
options, Non supported condition ${condition.type}, output);
}
/**
-
Adds a value inside of a node.
-
@param {!options} options Dictionary of configurations.
-
@param {!value} value Value to display.
-
@param {!output} output Output display accumulator.
*/
function display_value(options, value, output) {
if (value.type === ‘PROBABILITY’) {
const left_margin = 0;
const right_margin = 50;
const plot_width = options.node_x_size - options.node_padding * 2 -
left_margin - right_margin;let cusum = Array.from(d3.cumsum(value.distribution));
cusum.unshift(0);
const distribution_plot = output.content.append(‘g’).attr(
‘transform’,translate(0,${output.vertical_offset + 0.5}));distribution_plot.selectAll(‘rect’)
.data(value.distribution)
.join(‘rect’)
.attr(‘height’, 10)
.attr(
‘x’,
(d, i) =>
(cusum[i] * plot_width + left_margin + options.node_padding))
.attr(‘width’, (d, i) => d * plot_width)
.style(‘fill’, (d, i) => d3.schemeSet1[i]);const num_examples =
output.content.append(‘g’)
.attr(‘transform’,translate(0,${output.vertical_offset}))
.append(‘text’)
.attr(‘x’, options.node_x_size - options.node_padding)
.attr(‘alignment-baseline’, ‘hanging’)
.attr(‘text-anchor’, ‘end’)
.text((${value.num_examples}));const distribution_details = d3.create(‘ul’);
distribution_details.selectAll(‘li’)
.data(value.distribution)
.join(‘li’)
.append(‘span’)
.text(
(d, i) =>
‘class ’ + i + ‘: ’ + d3.format(’.3%’)(value.distribution[i]));add_tooltip(options, distribution_plot, () => distribution_details.html());
add_tooltip(options, num_examples, () => ‘Number of examples’);output.vertical_offset += 10;
return;
}
if (value.type === ‘REGRESSION’) {
display_node_text(
options,
‘value: ’ + d3.format(‘r’)(value.value) + ( +
d3.format(’.6’)(value.num_examples) + ),
output);
return;
}
display_node_text(options, Non supported value ${value.type}, output);
}
/**
- Adds an explanation inside of a node.
- @param {!options} options Dictionary of configurations.
- @param {!explanation} explanation Explanation to display.
- @param {!output} output Output display accumulator.
*/
function display_explanation(options, explanation, output) {
// Margin before the explanation.
output.vertical_offset += 10;
display_node_text(
options, Non supported explanation ${explanation.type}, output);
}
/**
- Draw the edges of the tree.
- @param {!options} options Dictionary of configurations.
- @param {!graph} graph D3 search handle containing the graph.
- @param {!tree_struct} tree_struct Structure of the tree (node placement,
-
data, etc.).
*/
function display_edges(options, graph, tree_struct) {
// Draw an edge between a parent and a child node with a bezier.
function draw_single_edge(d) {
return ‘M’ + (d.source.y + options.node_x_size) + ‘,’ + d.source.x + ’ C’ +
(d.source.y + options.node_x_size + options.edge_rounding) + ‘,’ +
d.source.x + ’ ’ + (d.target.y - options.edge_rounding) + ‘,’ +
d.target.x + ’ ’ + d.target.y + ‘,’ + d.target.x;
}
graph.append(‘g’)
.attr(‘fill’, ‘none’)
.attr(‘stroke-width’, 1.2)
.selectAll(‘path’)
.data(tree_struct.links())
.join(‘path’)
.attr(‘d’, draw_single_edge)
.attr(
‘stroke’, d => (d.target === d.source.children[0]) ? ‘#0F0’ : ‘#F00’);
}
display_tree({“margin”: 10, “node_x_size”: 160, “node_y_size”: 28, “node_x_offset”: 180, “node_y_offset”: 33, “font_size”: 10, “edge_rounding”: 20, “node_padding”: 2, “show_plot_bounding_box”: false}, {“value”: {“type”: “PROBABILITY”, “distribution”: [0.47093023255813954, 0.19476744186046513, 0.33430232558139533], “num_examples”: 344.0}, “condition”: {“type”: “NUMERICAL_IS_HIGHER_THAN”, “attribute”: “bill_length_mm”, “threshold”: 43.25}, “children”: [{“value”: {“type”: “PROBABILITY”, “distribution”: [0.005847953216374269, 0.3567251461988304, 0.6374269005847953], “num_examples”: 171.0}, “condition”: {“type”: “CATEGORICAL_IS_IN”, “attribute”: “island”, “mask”: [“Biscoe”]}, “children”: [{“value”: {“type”: “PROBABILITY”, “distribution”: [0.00909090909090909, 0.0, 0.990909090909091], “num_examples”: 110.0}, “condition”: {“type”: “NUMERICAL_IS_HIGHER_THAN”, “attribute”: “bill_depth_mm”, “threshold”: 17.225584030151367}, “children”: [{“value”: {“type”: “PROBABILITY”, “distribution”: [0.16666666666666666, 0.0, 0.8333333333333334], “num_examples”: 6.0}}, {“value”: {“type”: “PROBABILITY”, “distribution”: [0.0, 0.0, 1.0], “num_examples”: 104.0}}]}, {“value”: {“type”: “PROBABILITY”, “distribution”: [0.0, 1.0, 0.0], “num_examples”: 61.0}}]}, {“value”: {“type”: “PROBABILITY”, “distribution”: [0.930635838150289, 0.03468208092485549, 0.03468208092485549], “num_examples”: 173.0}, “condition”: {“type”: “NUMERICAL_IS_HIGHER_THAN”, “attribute”: “bill_depth_mm”, “threshold”: 15.100000381469727}, “children”: [{“value”: {“type”: “PROBABILITY”, “distribution”: [0.9640718562874252, 0.03592814371257485, 0.0], “num_examples”: 167.0}, “condition”: {“type”: “NUMERICAL_IS_HIGHER_THAN”, “attribute”: “flipper_length_mm”, “threshold”: 187.5}, “children”: [{“value”: {“type”: “PROBABILITY”, “distribution”: [1.0, 0.0, 0.0], “num_examples”: 104.0}}, {“value”: {“type”: “PROBABILITY”, “distribution”: [0.9047619047619048, 0.09523809523809523, 0.0], “num_examples”: 63.0}, “condition”: {“type”: “NUMERICAL_IS_HIGHER_THAN”, “attribute”: “bill_length_mm”, “threshold”: 42.30000305175781}}]}, {“value”: {“type”: “PROBABILITY”, “distribution”: [0.0, 0.0, 1.0], “num_examples”: 6.0}}]}]}, “#tree_plot_05707b35c4f748738efd3da21ab9197f”)
检查模型结构
模型结构和元数据可以通过make_inspector()创建的inspector来获取。
**注意:**根据学习算法和超参数的不同,inspector将暴露不同的专门属性。例如,winner_take_all字段是随机森林模型特有的。
# 创建一个模型检查器对象,用于检查模型的性能和质量
inspector = model.make_inspector()
对于我们的模型,可用的检查员字段有:
# 使用列表推导式,遍历inspector模块中的所有属性
# 过滤掉以"_"开头的属性
fields = [field for field in dir(inspector) if not field.startswith("_")]
['MODEL_NAME',
'dataspec',
'evaluation',
'export_to_tensorboard',
'extract_all_trees',
'extract_tree',
'features',
'header',
'iterate_on_nodes',
'label',
'label_classes',
'metadata',
'model_type',
'num_trees',
'objective',
'specialized_header',
'task',
'training_logs',
'tuning_logs',
'variable_importances',
'winner_take_all_inference']
记得查看API参考或使用?查看内置文档。
?inspector.model_type
一些模型元数据:
# 打印模型类型
print("Model type:", inspector.model_type())
# 打印模型中树的数量
print("Number of trees:", inspector.num_trees())
# 打印模型的目标函数
print("Objective:", inspector.objective())
# 打印模型的输入特征
print("Input features:", inspector.features())
Model type: RANDOM_FOREST
Number of trees: 300
Objective: Classification(label=__LABEL, class=None, num_classes=3)
Input features: ["bill_depth_mm" (1; #0), "bill_length_mm" (1; #1), "body_mass_g" (1; #2), "flipper_length_mm" (1; #3), "island" (4; #4), "sex" (4; #5), "year" (1; #6)]
evaluate()是在训练期间计算的模型评估。用于此评估的数据集取决于算法。例如,它可以是验证数据集或袋外数据集。
**注意:**虽然在训练期间计算,但evaluate()从未对训练数据集进行评估。
# 创建一个名为inspector的对象
inspector = Inspector()
# 调用inspector对象的evaluation()方法
inspector.evaluation()
Evaluation(num_examples=344, accuracy=0.9767441860465116, loss=0.06782230959804512, rmse=None, ndcg=None, aucs=None, auuc=None, qini=None)
变量重要性如下:
The variable importances are:
# 打印可用的变量重要性
print(f"Available variable importances:")
# 遍历变量重要性字典的键,并打印出来
for importance in inspector.variable_importances().keys():
print("\t", importance)
Available variable importances:
MEAN_DECREASE_IN_AP_1_VS_OTHERS
MEAN_DECREASE_IN_PRAUC_3_VS_OTHERS
SUM_SCORE
MEAN_DECREASE_IN_PRAUC_1_VS_OTHERS
MEAN_DECREASE_IN_ACCURACY
MEAN_DECREASE_IN_AUC_1_VS_OTHERS
MEAN_DECREASE_IN_AP_3_VS_OTHERS
NUM_AS_ROOT
MEAN_DECREASE_IN_AP_2_VS_OTHERS
MEAN_DECREASE_IN_AUC_2_VS_OTHERS
MEAN_MIN_DEPTH
MEAN_DECREASE_IN_AUC_3_VS_OTHERS
NUM_NODES
MEAN_DECREASE_IN_PRAUC_2_VS_OTHERS
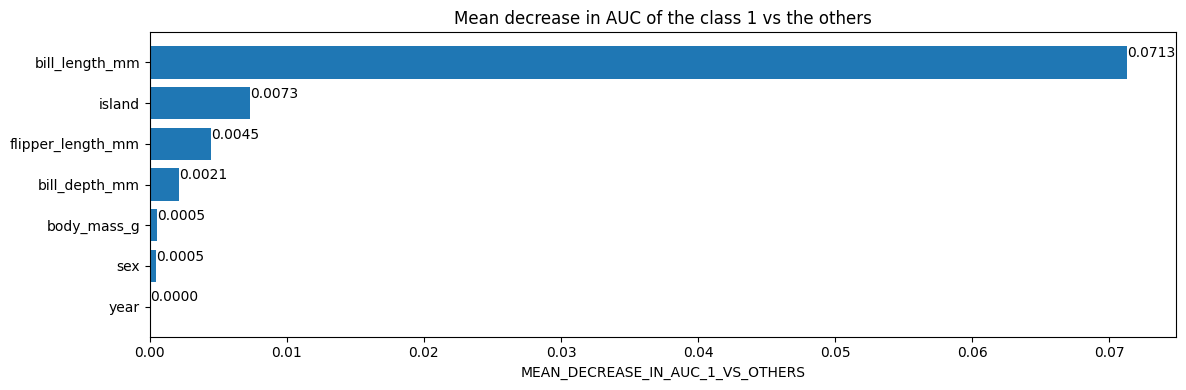
不同的变量重要性具有不同的语义。例如,具有平均减少auc为0.05的特征意味着从训练数据集中移除该特征会使AUC降低/受损5%。
# 获取类别1与其他类别之间的AUC的平均减少量
mean_decrease_in_auc_1_vs_others = inspector.variable_importances()["MEAN_DECREASE_IN_AUC_1_VS_OTHERS"]
[("bill_length_mm" (1; #1), 0.0713061951754389),
("island" (4; #4), 0.007298519736842035),
("flipper_length_mm" (1; #3), 0.004505893640351366),
("bill_depth_mm" (1; #0), 0.0021244517543865804),
("body_mass_g" (1; #2), 0.0005482456140351033),
("sex" (4; #5), 0.00047971491228060437),
("year" (1; #6), 0.0)]
绘制使用Matplotlib的检查器中的变量重要性
import matplotlib.pyplot as plt
plt.figure(figsize=(12, 4)) # 创建一个大小为12x4的图形
# 平均AUC下降值(class 1相对于其他类别)
variable_importance_metric = "MEAN_DECREASE_IN_AUC_1_VS_OTHERS"
variable_importances = inspector.variable_importances()[variable_importance_metric]
# 提取特征名称和重要性值
#
# `variable_importances` 是一个包含<特征, 重要性>元组的列表
feature_names = [vi[0].name for vi in variable_importances] # 提取特征名称
feature_importances = [vi[1] for vi in variable_importances] # 提取重要性值
# 特征按重要性值降序排列
feature_ranks = range(len(feature_names))
bar = plt.barh(feature_ranks, feature_importances, label=[str(x) for x in feature_ranks]) # 创建水平条形图
plt.yticks(feature_ranks, feature_names) # 设置y轴刻度为特征名称
plt.gca().invert_yaxis() # 反转y轴刻度顺序,使重要性高的特征在上方
# TODO: 当可用时,替换为 "plt.bar_label()"
# 使用值标记每个条形图
for importance, patch in zip(feature_importances, bar.patches):
plt.text(patch.get_x() + patch.get_width(), patch.get_y(), f"{importance:.4f}", va="top")
plt.xlabel(variable_importance_metric) # 设置x轴标签为重要性度量
plt.title("Mean decrease in AUC of the class 1 vs the others") # 设置图形标题
plt.tight_layout() # 调整图形布局,以防止标签重叠
plt.show() # 显示图形
最后,访问实际的树结构:
# 从inspector对象中提取树的信息
# 参数tree_idx表示要提取的树的索引,这里为0表示提取第一棵树的信息
inspector.extract_tree(tree_idx=0)
Tree(root=NonLeafNode(condition=(bill_length_mm >= 43.25; miss=True, score=0.5482327342033386), pos_child=NonLeafNode(condition=(island in ['Biscoe']; miss=True, score=0.6515106558799744), pos_child=NonLeafNode(condition=(bill_depth_mm >= 17.225584030151367; miss=False, score=0.027205035090446472), pos_child=LeafNode(value=ProbabilityValue([0.16666666666666666, 0.0, 0.8333333333333334],n=6.0), idx=7), neg_child=LeafNode(value=ProbabilityValue([0.0, 0.0, 1.0],n=104.0), idx=6), value=ProbabilityValue([0.00909090909090909, 0.0, 0.990909090909091],n=110.0)), neg_child=LeafNode(value=ProbabilityValue([0.0, 1.0, 0.0],n=61.0), idx=5), value=ProbabilityValue([0.005847953216374269, 0.3567251461988304, 0.6374269005847953],n=171.0)), neg_child=NonLeafNode(condition=(bill_depth_mm >= 15.100000381469727; miss=True, score=0.150658518075943), pos_child=NonLeafNode(condition=(flipper_length_mm >= 187.5; miss=True, score=0.036139510571956635), pos_child=LeafNode(value=ProbabilityValue([1.0, 0.0, 0.0],n=104.0), idx=4), neg_child=NonLeafNode(condition=(bill_length_mm >= 42.30000305175781; miss=True, score=0.23430533707141876), pos_child=LeafNode(value=ProbabilityValue([0.0, 1.0, 0.0],n=5.0), idx=3), neg_child=NonLeafNode(condition=(bill_length_mm >= 40.55000305175781; miss=True, score=0.043961383402347565), pos_child=LeafNode(value=ProbabilityValue([0.8, 0.2, 0.0],n=5.0), idx=2), neg_child=LeafNode(value=ProbabilityValue([1.0, 0.0, 0.0],n=53.0), idx=1), value=ProbabilityValue([0.9827586206896551, 0.017241379310344827, 0.0],n=58.0)), value=ProbabilityValue([0.9047619047619048, 0.09523809523809523, 0.0],n=63.0)), value=ProbabilityValue([0.9640718562874252, 0.03592814371257485, 0.0],n=167.0)), neg_child=LeafNode(value=ProbabilityValue([0.0, 0.0, 1.0],n=6.0), idx=0), value=ProbabilityValue([0.930635838150289, 0.03468208092485549, 0.03468208092485549],n=173.0)), value=ProbabilityValue([0.47093023255813954, 0.19476744186046513, 0.33430232558139533],n=344.0)), label_classes=None)
提取树并不高效。如果速度很重要,可以使用iterate_on_nodes()方法来进行模型检查。这个方法是对模型的所有节点进行深度优先的前序遍历迭代器。
注意:extract_tree()是使用iterate_on_nodes()实现的。
以下示例计算每个特征被使用的次数(这是一种结构变量重要性的指标):
# 创建一个默认字典number_of_use,用于记录每个特征在其条件中被使用的次数
number_of_use = collections.defaultdict(lambda: 0)
# 对所有节点进行深度优先的前序遍历
for node_iter in inspector.iterate_on_nodes():
# 如果节点不是叶节点,则跳过
if not isinstance(node_iter.node, tfdf.py_tree.node.NonLeafNode):
continue
# 遍历节点条件中使用的所有特征
# 默认情况下,模型是"oblique"的,即每个节点测试一个特征
for feature in node_iter.node.condition.features():
# 特征在使用次数上加1
number_of_use[feature] += 1
# 打印每个特征的条件节点数
print("Number of condition nodes per features:")
for feature, count in number_of_use.items():
print("\t", feature.name, ":", count)
Number of condition nodes per features:
bill_length_mm : 778
bill_depth_mm : 463
flipper_length_mm : 414
island : 342
body_mass_g : 338
year : 19
sex : 36
手动创建模型
在本节中,您将手动创建一个小的随机森林模型。为了使其更加简单,该模型只包含一个简单的树:
3个标签类别:红色、蓝色和绿色。
2个特征:f1(数值型)和f2(字符串分类型)
f1>=1.5
├─(正)─ f2在["猫","狗"]中
│ ├─(正)─ 值:[0.8, 0.1, 0.1]
│ └─(负)─ 值:[0.1, 0.8, 0.1]
└─(负)─ 值:[0.1, 0.1, 0.8]
# 创建模型构建器
builder = tfdf.builder.RandomForestBuilder(
path="/tmp/manual_model", # 指定模型保存的路径
objective=tfdf.py_tree.objective.ClassificationObjective(
label="color", # 指定目标变量为"color"
classes=["red", "blue", "green"])) # 指定目标变量的类别为["red", "blue", "green"]
每棵树都逐个添加。
注意: 树对象(tfdf.py_tree.tree.Tree)与前一节中extract_tree()返回的树对象相同。
# 导入所需的模块和类
Tree = tfdf.py_tree.tree.Tree # 树结构
SimpleColumnSpec = tfdf.py_tree.dataspec.SimpleColumnSpec # 列规范
ColumnType = tfdf.py_tree.dataspec.ColumnType # 列类型
NonLeafNode = tfdf.py_tree.node.NonLeafNode # 非叶节点
LeafNode = tfdf.py_tree.node.LeafNode # 叶节点
NumericalHigherThanCondition = tfdf.py_tree.condition.NumericalHigherThanCondition # 数值大于条件
CategoricalIsInCondition = tfdf.py_tree.condition.CategoricalIsInCondition # 类别在条件
ProbabilityValue = tfdf.py_tree.value.ProbabilityValue # 概率值
# 创建树结构并添加到builder中
builder.add_tree(
Tree(
NonLeafNode(
condition=NumericalHigherThanCondition(
feature=SimpleColumnSpec(name="f1", type=ColumnType.NUMERICAL), # 数值特征"f1"
threshold=1.5, # 阈值为1.5
missing_evaluation=False), # 不考虑缺失值
pos_child=NonLeafNode(
condition=CategoricalIsInCondition(
feature=SimpleColumnSpec(name="f2",type=ColumnType.CATEGORICAL), # 类别特征"f2"
mask=["cat", "dog"], # 类别为"cat"或"dog"
missing_evaluation=False), # 不考虑缺失值
pos_child=LeafNode(value=ProbabilityValue(probability=[0.8, 0.1, 0.1], num_examples=10)), # 正向子节点为叶节点,概率值为[0.8, 0.1, 0.1],样本数为10
neg_child=LeafNode(value=ProbabilityValue(probability=[0.1, 0.8, 0.1], num_examples=20))), # 负向子节点为叶节点,概率值为[0.1, 0.8, 0.1],样本数为20
neg_child=LeafNode(value=ProbabilityValue(probability=[0.1, 0.1, 0.8], num_examples=30))))) # 负向子节点为叶节点,概率值为[0.1, 0.1, 0.8],样本数为30
结束树写作
# 关闭builder对象
builder.close()
[INFO 2022-12-14T12:25:00.790486355+00:00 kernel.cc:1175] Loading model from path /tmp/manual_model/tmp/ with prefix e09a067144bc479b
[INFO 2022-12-14T12:25:00.790802259+00:00 decision_forest.cc:640] Model loaded with 1 root(s), 5 node(s), and 2 input feature(s).
[INFO 2022-12-14T12:25:00.790878962+00:00 kernel.cc:1021] Use fast generic engine
WARNING:absl:Found untraced functions such as call_get_leaves, _update_step_xla while saving (showing 2 of 2). These functions will not be directly callable after loading.
INFO:tensorflow:Assets written to: /tmp/manual_model/assets
INFO:tensorflow:Assets written to: /tmp/manual_model/assets
现在您可以将该模型作为常规的keras模型打开,并进行预测:
# 加载预训练模型
manual_model = tf.keras.models.load_model("/tmp/manual_model")
[INFO 2022-12-14T12:25:01.436506097+00:00 kernel.cc:1175] Loading model from path /tmp/manual_model/assets/ with prefix e09a067144bc479b
[INFO 2022-12-14T12:25:01.436871761+00:00 decision_forest.cc:640] Model loaded with 1 root(s), 5 node(s), and 2 input feature(s).
[INFO 2022-12-14T12:25:01.436909696+00:00 kernel.cc:1021] Use fast generic engine
# 创建一个tf.data.Dataset对象,从给定的张量中切片得到数据集
# 数据集包含两个特征"f1"和"f2",分别是浮点数和字符串类型
# 数据集中的每个样本是一个字典,包含"f1"和"f2"两个键
# 样本数据为:
# "f1": [1.0, 2.0, 3.0]
# "f2": ["cat", "cat", "bird"]
# 使用batch(2)方法将数据集划分为大小为2的批次
examples = tf.data.Dataset.from_tensor_slices({
"f1": [1.0, 2.0, 3.0],
"f2": ["cat", "cat", "bird"]
}).batch(2)
# 使用manual_model对examples进行预测
predictions = manual_model.predict(examples)
# 打印预测结果
print("predictions:\n", predictions)
1/2 [==============>...............] - ETA: 0s
2/2 [==============================] - 0s 2ms/step
predictions:
[[0.1 0.1 0.8]
[0.8 0.1 0.1]
[0.1 0.8 0.1]]
访问结构:
注意: 由于模型是序列化和反序列化的,您需要使用一种替代但等效的形式。
# 代码注释
# 获取yggdrasil模型路径
yggdrasil_model_path = manual_model.yggdrasil_model_path_tensor().numpy().decode("utf-8")
print("yggdrasil_model_path:",yggdrasil_model_path)
# 创建一个模型检查器,用于检查模型的输入特征
inspector = tfdf.inspector.make_inspector(yggdrasil_model_path)
print("Input features:", inspector.features())
yggdrasil_model_path: /tmp/manual_model/assets/
Input features: ["f1" (1; #1), "f2" (4; #2)]
当然,您可以手动绘制这个构建的模型:
# 导入tfdf库中的plot_model_in_colab函数
import tensorflow_decision_forests as tfdf
# 使用plot_model_in_colab函数绘制manual_model模型的结构图
tfdf.model_plotter.plot_model_in_colab(manual_model)
/**
- Plotting of decision trees generated by TF-DF.
- A tree is a recursive structure of node objects.
- A node contains one or more of the following components:
-
- A value: Representing the output of the node. If the node is not a leaf,
-
the value is only present for analysis i.e. it is not used for -
predictions. -
- A condition : For non-leaf nodes, the condition (also known as split)
-
defines a binary test to branch to the positive or negative child. -
- An explanation: Generally a plot showing the relation between the label
-
and the condition to give insights about the effect of the condition. -
- Two children : For non-leaf nodes, the children nodes. The first
-
children (i.e. "node.children[0]") is the negative children (drawn in -
red). The second children is the positive one (drawn in green).
*/
/**
- Plots a single decision tree into a DOM element.
- @param {!options} options Dictionary of configurations.
- @param {!tree} raw_tree Recursive tree structure.
- @param {string} canvas_id Id of the output dom element.
*/
function display_tree(options, raw_tree, canvas_id) {
console.log(options);
// Determine the node placement.
const tree_struct = d3.tree().nodeSize(
[options.node_y_offset, options.node_x_offset])(d3.hierarchy(raw_tree));
// Boundaries of the node placement.
let x_min = Infinity;
let x_max = -x_min;
let y_min = Infinity;
let y_max = -x_min;
tree_struct.each(d => {
if (d.x > x_max) x_max = d.x;
if (d.x < x_min) x_min = d.x;
if (d.y > y_max) y_max = d.y;
if (d.y < y_min) y_min = d.y;
});
// Size of the plot.
const width = y_max - y_min + options.node_x_size + options.margin * 2;
const height = x_max - x_min + options.node_y_size + options.margin * 2 +
options.node_y_offset - options.node_y_size;
const plot = d3.select(canvas_id);
// Tool tip
options.tooltip = plot.append(‘div’)
.attr(‘width’, 100)
.attr(‘height’, 100)
.style(‘padding’, ‘4px’)
.style(‘background’, ‘#fff’)
.style(‘box-shadow’, ‘4px 4px 0px rgba(0,0,0,0.1)’)
.style(‘border’, ‘1px solid black’)
.style(‘font-family’, ‘sans-serif’)
.style(‘font-size’, options.font_size)
.style(‘position’, ‘absolute’)
.style(‘z-index’, ‘10’)
.attr(‘pointer-events’, ‘none’)
.style(‘display’, ‘none’);
// Create canvas
const svg = plot.append(‘svg’).attr(‘width’, width).attr(‘height’, height);
const graph =
svg.style(‘overflow’, ‘visible’)
.append(‘g’)
.attr(‘font-family’, ‘sans-serif’)
.attr(‘font-size’, options.font_size)
.attr(
‘transform’,
() => translate(${options.margin},${ - x_min + options.node_y_offset / 2 + options.margin}));
// Plot bounding box.
if (options.show_plot_bounding_box) {
svg.append(‘rect’)
.attr(‘width’, width)
.attr(‘height’, height)
.attr(‘fill’, ‘none’)
.attr(‘stroke-width’, 1.0)
.attr(‘stroke’, ‘black’);
}
// Draw the edges.
display_edges(options, graph, tree_struct);
// Draw the nodes.
display_nodes(options, graph, tree_struct);
}
/**
- Draw the nodes of the tree.
- @param {!options} options Dictionary of configurations.
- @param {!graph} graph D3 search handle containing the graph.
- @param {!tree_struct} tree_struct Structure of the tree (node placement,
-
data, etc.).
*/
function display_nodes(options, graph, tree_struct) {
const nodes = graph.append(‘g’)
.selectAll(‘g’)
.data(tree_struct.descendants())
.join(‘g’)
.attr(‘transform’, d => translate(${d.y},${d.x}));
nodes.append(‘rect’)
.attr(‘x’, 0.5)
.attr(‘y’, 0.5)
.attr(‘width’, options.node_x_size)
.attr(‘height’, options.node_y_size)
.attr(‘stroke’, ‘lightgrey’)
.attr(‘stroke-width’, 1)
.attr(‘fill’, ‘white’)
.attr(‘y’, -options.node_y_size / 2);
// Brackets on the right of condition nodes without children.
non_leaf_node_without_children =
nodes.filter(node => node.data.condition != null && node.children == null)
.append(‘g’)
.attr(‘transform’, translate(${options.node_x_size},0));
non_leaf_node_without_children.append(‘path’)
.attr(‘d’, ‘M0,0 C 10,0 0,10 10,10’)
.attr(‘fill’, ‘none’)
.attr(‘stroke-width’, 1.0)
.attr(‘stroke’, ‘#F00’);
non_leaf_node_without_children.append(‘path’)
.attr(‘d’, ‘M0,0 C 10,0 0,-10 10,-10’)
.attr(‘fill’, ‘none’)
.attr(‘stroke-width’, 1.0)
.attr(‘stroke’, ‘#0F0’);
const node_content = nodes.append(‘g’).attr(
‘transform’,
translate(0,${options.node_padding - options.node_y_size / 2}));
node_content.append(node => create_node_element(options, node));
}
/**
- Creates the D3 content for a single node.
- @param {!options} options Dictionary of configurations.
- @param {!node} node Node to draw.
- @return {!d3} D3 content.
*/
function create_node_element(options, node) {
// Output accumulator.
let output = {
// Content to draw.
content: d3.create(‘svg:g’),
// Vertical offset to the next element to draw.
vertical_offset: 0
};
// Conditions.
if (node.data.condition != null) {
display_condition(options, node.data.condition, output);
}
// Values.
if (node.data.value != null) {
display_value(options, node.data.value, output);
}
// Explanations.
if (node.data.explanation != null) {
display_explanation(options, node.data.explanation, output);
}
return output.content.node();
}
/**
- Adds a single line of text inside of a node.
- @param {!options} options Dictionary of configurations.
- @param {string} text Text to display.
- @param {!output} output Output display accumulator.
*/
function display_node_text(options, text, output) {
output.content.append(‘text’)
.attr(‘x’, options.node_padding)
.attr(‘y’, output.vertical_offset)
.attr(‘alignment-baseline’, ‘hanging’)
.text(text);
output.vertical_offset += 10;
}
/**
- Adds a single line of text inside of a node with a tooltip.
- @param {!options} options Dictionary of configurations.
- @param {string} text Text to display.
- @param {string} tooltip Text in the Tooltip.
- @param {!output} output Output display accumulator.
*/
function display_node_text_with_tooltip(options, text, tooltip, output) {
const item = output.content.append(‘text’)
.attr(‘x’, options.node_padding)
.attr(‘alignment-baseline’, ‘hanging’)
.text(text);
add_tooltip(options, item, () => tooltip);
output.vertical_offset += 10;
}
/**
- Adds a tooltip to a dom element.
- @param {!options} options Dictionary of configurations.
- @param {!dom} target Dom element to equip with a tooltip.
- @param {!func} get_content Generates the html content of the tooltip.
*/
function add_tooltip(options, target, get_content) {
function show(d) {
options.tooltip.style(‘display’, ‘block’);
options.tooltip.html(get_content());
}
function hide(d) {
options.tooltip.style(‘display’, ‘none’);
}
function move(d) {
options.tooltip.style(‘display’, ‘block’);
options.tooltip.style(‘left’, (d.pageX + 5) + ‘px’);
options.tooltip.style(‘top’, d.pageY + ‘px’);
}
target.on(‘mouseover’, show);
target.on(‘mouseout’, hide);
target.on(‘mousemove’, move);
}
/**
- Adds a condition inside of a node.
- @param {!options} options Dictionary of configurations.
- @param {!condition} condition Condition to display.
- @param {!output} output Output display accumulator.
*/
function display_condition(options, condition, output) {
threshold_format = d3.format(‘r’);
if (condition.type === ‘IS_MISSING’) {
display_node_text(options, ${condition.attribute} is missing, output);
return;
}
if (condition.type === ‘IS_TRUE’) {
display_node_text(options, ${condition.attribute} is true, output);
return;
}
if (condition.type === ‘NUMERICAL_IS_HIGHER_THAN’) {
format = d3.format(‘r’);
display_node_text(
options,
${condition.attribute} >= ${threshold_format(condition.threshold)},
output);
return;
}
if (condition.type === ‘CATEGORICAL_IS_IN’) {
display_node_text_with_tooltip(
options, ${condition.attribute} in [...],
${condition.attribute} in [${condition.mask}], output);
return;
}
if (condition.type === ‘CATEGORICAL_SET_CONTAINS’) {
display_node_text_with_tooltip(
options, ${condition.attribute} intersect [...],
${condition.attribute} intersect [${condition.mask}], output);
return;
}
if (condition.type === ‘NUMERICAL_SPARSE_OBLIQUE’) {
display_node_text_with_tooltip(
options, Sparse oblique split...,
[${condition.attributes}]*[${condition.weights}]>=${ threshold_format(condition.threshold)},
output);
return;
}
display_node_text(
options, Non supported condition ${condition.type}, output);
}
/**
-
Adds a value inside of a node.
-
@param {!options} options Dictionary of configurations.
-
@param {!value} value Value to display.
-
@param {!output} output Output display accumulator.
*/
function display_value(options, value, output) {
if (value.type === ‘PROBABILITY’) {
const left_margin = 0;
const right_margin = 50;
const plot_width = options.node_x_size - options.node_padding * 2 -
left_margin - right_margin;let cusum = Array.from(d3.cumsum(value.distribution));
cusum.unshift(0);
const distribution_plot = output.content.append(‘g’).attr(
‘transform’,translate(0,${output.vertical_offset + 0.5}));distribution_plot.selectAll(‘rect’)
.data(value.distribution)
.join(‘rect’)
.attr(‘height’, 10)
.attr(
‘x’,
(d, i) =>
(cusum[i] * plot_width + left_margin + options.node_padding))
.attr(‘width’, (d, i) => d * plot_width)
.style(‘fill’, (d, i) => d3.schemeSet1[i]);const num_examples =
output.content.append(‘g’)
.attr(‘transform’,translate(0,${output.vertical_offset}))
.append(‘text’)
.attr(‘x’, options.node_x_size - options.node_padding)
.attr(‘alignment-baseline’, ‘hanging’)
.attr(‘text-anchor’, ‘end’)
.text((${value.num_examples}));const distribution_details = d3.create(‘ul’);
distribution_details.selectAll(‘li’)
.data(value.distribution)
.join(‘li’)
.append(‘span’)
.text(
(d, i) =>
‘class ’ + i + ‘: ’ + d3.format(’.3%’)(value.distribution[i]));add_tooltip(options, distribution_plot, () => distribution_details.html());
add_tooltip(options, num_examples, () => ‘Number of examples’);output.vertical_offset += 10;
return;
}
if (value.type === ‘REGRESSION’) {
display_node_text(
options,
‘value: ’ + d3.format(‘r’)(value.value) + ( +
d3.format(’.6’)(value.num_examples) + ),
output);
return;
}
display_node_text(options, Non supported value ${value.type}, output);
}
/**
- Adds an explanation inside of a node.
- @param {!options} options Dictionary of configurations.
- @param {!explanation} explanation Explanation to display.
- @param {!output} output Output display accumulator.
*/
function display_explanation(options, explanation, output) {
// Margin before the explanation.
output.vertical_offset += 10;
display_node_text(
options, Non supported explanation ${explanation.type}, output);
}
/**
- Draw the edges of the tree.
- @param {!options} options Dictionary of configurations.
- @param {!graph} graph D3 search handle containing the graph.
- @param {!tree_struct} tree_struct Structure of the tree (node placement,
-
data, etc.).
*/
function display_edges(options, graph, tree_struct) {
// Draw an edge between a parent and a child node with a bezier.
function draw_single_edge(d) {
return ‘M’ + (d.source.y + options.node_x_size) + ‘,’ + d.source.x + ’ C’ +
(d.source.y + options.node_x_size + options.edge_rounding) + ‘,’ +
d.source.x + ’ ’ + (d.target.y - options.edge_rounding) + ‘,’ +
d.target.x + ’ ’ + d.target.y + ‘,’ + d.target.x;
}
graph.append(‘g’)
.attr(‘fill’, ‘none’)
.attr(‘stroke-width’, 1.2)
.selectAll(‘path’)
.data(tree_struct.links())
.join(‘path’)
.attr(‘d’, draw_single_edge)
.attr(
‘stroke’, d => (d.target === d.source.children[0]) ? ‘#0F0’ : ‘#F00’);
}
display_tree({“margin”: 10, “node_x_size”: 160, “node_y_size”: 28, “node_x_offset”: 180, “node_y_offset”: 33, “font_size”: 10, “edge_rounding”: 20, “node_padding”: 2, “show_plot_bounding_box”: false, “labels”: “[“red”, “blue”, “green”]”}, {“condition”: {“type”: “NUMERICAL_IS_HIGHER_THAN”, “attribute”: “f1”, “threshold”: 1.5}, “children”: [{“condition”: {“type”: “CATEGORICAL_IS_IN”, “attribute”: “f2”, “mask”: [“cat”, “dog”]}, “children”: [{“value”: {“type”: “PROBABILITY”, “distribution”: [0.8, 0.1, 0.1], “num_examples”: 10.0}}, {“value”: {“type”: “PROBABILITY”, “distribution”: [0.1, 0.8, 0.1], “num_examples”: 20.0}}]}, {“value”: {“type”: “PROBABILITY”, “distribution”: [0.1, 0.1, 0.8], “num_examples”: 30.0}}]}, “#tree_plot_34c8fb6cf7ca49eda845b971be7f0560”)
本文来自互联网用户投稿,该文观点仅代表作者本人,不代表本站立场。本站仅提供信息存储空间服务,不拥有所有权,不承担相关法律责任。 如若内容造成侵权/违法违规/事实不符,请联系我的编程经验分享网邮箱:chenni525@qq.com进行投诉反馈,一经查实,立即删除!