【bioinfo】收藏生信常用网址
文章目录
文件格式文档SAM/VCF
sam: https://samtools.github.io/hts-specs/SAMv1.pdf
vcf: http://samtools.github.io/hts-specs/VCFv4.3.pdf
工具手册bwa/samtools
bwa使用:http://bio-bwa.sourceforge.net/bwa.shtml
samtools使用:http://www.htslib.org/doc/samtools.html
samtools构建的索引.fai文件格式:https://www.htslib.org/doc/faidx.html
基因组统计学wiki
https://genome.sph.umich.edu/wiki/Variant_Normalization
比如:变异校正(Variant Normalization)查询
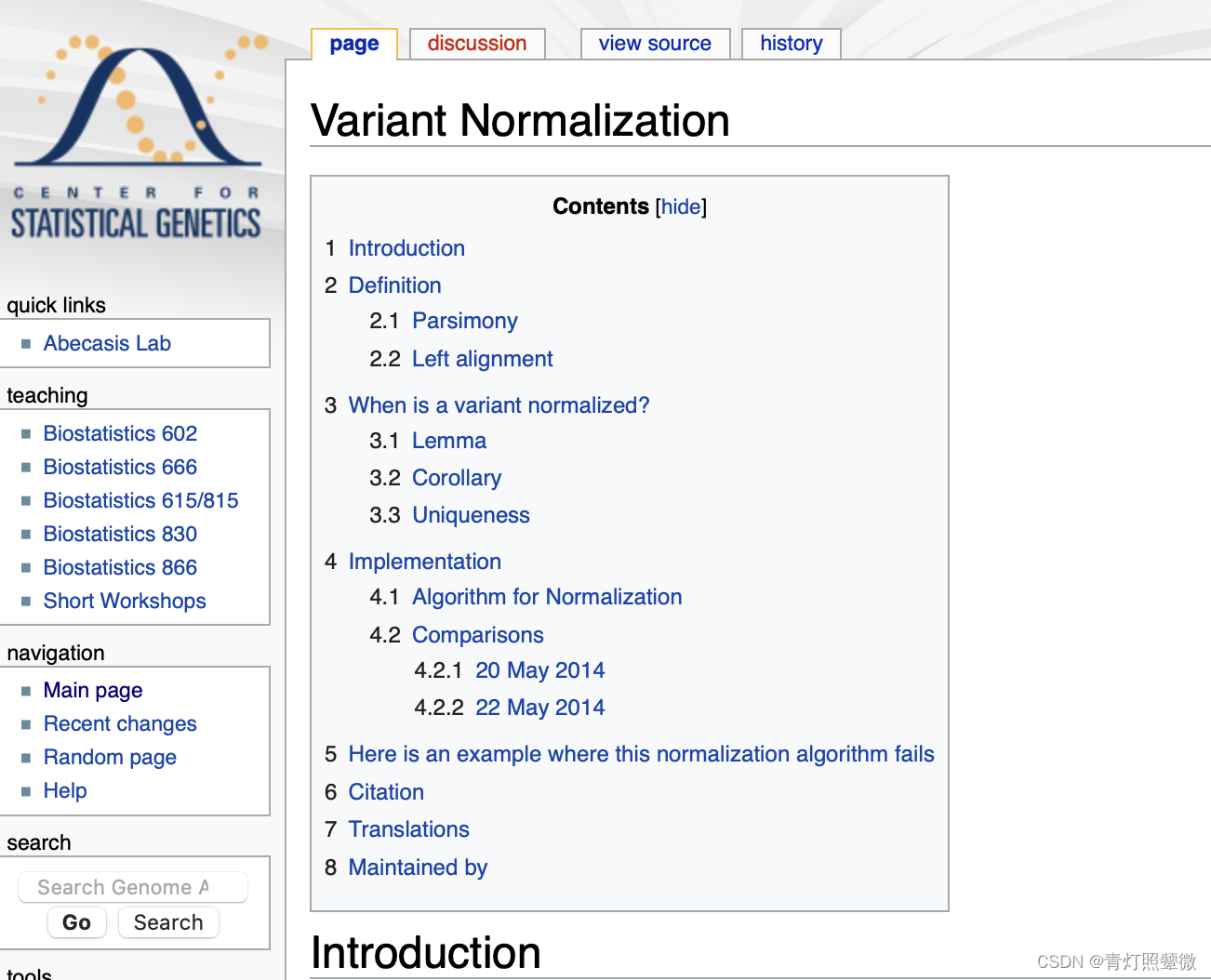
sam flag值查询
sam flag值对应信息: https://broadinstitute.github.io/picard/explain-flags.html
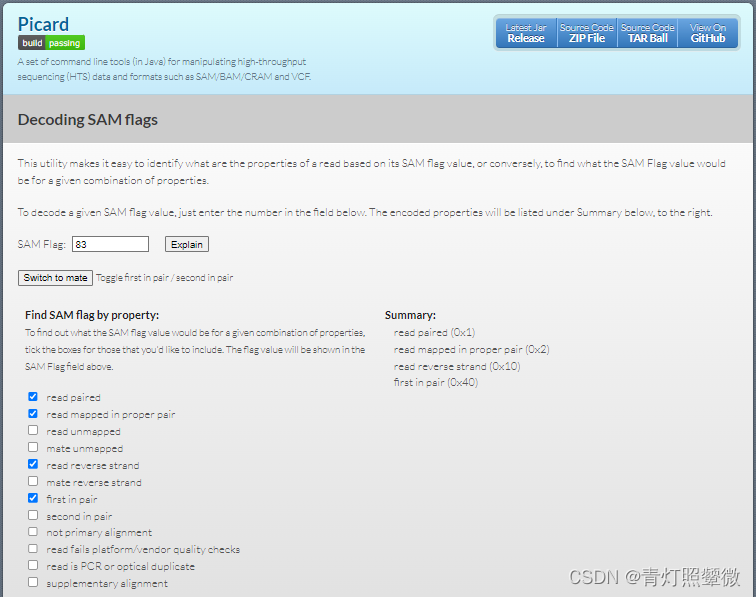
序列反向互补
http://www.bioinformatics.org/sms/rev_comp.html
TransVar 变异注释
在线工具:https://bioinformatics.mdanderson.org/transvar/
UCSC-blat在线比对
使用UCSC-blat进行序列比对:http://genome.ucsc.edu/cgi-bin/hgBlat
UCSC工具:https://genome.ucsc.edu/goldenPath/releaseLog.html#hg19
常用数据库 NCBI/nsembl/HGNC
NCBI:https://www.ncbi.nlm.nih.gov/

Ensembl:http://asia.ensembl.org/index.html

HGNC:https://www.genenames.org/about/guidelines/
论坛 biostars/SEQanswers
查询文献影响因子
https://www.pubmed.pro/home
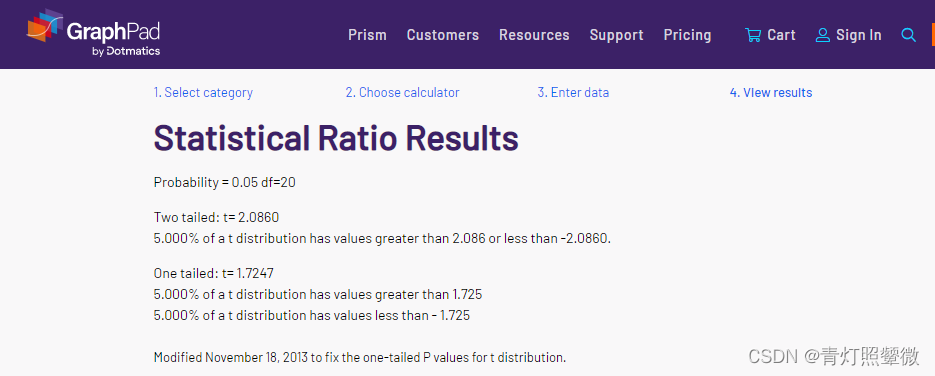
假设检验查询
https://www.graphpad.com/quickcalcs/statratio1/
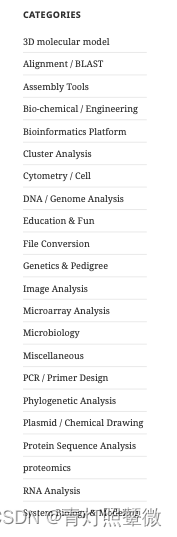
生信软件查询
https://mybiosoftware.com
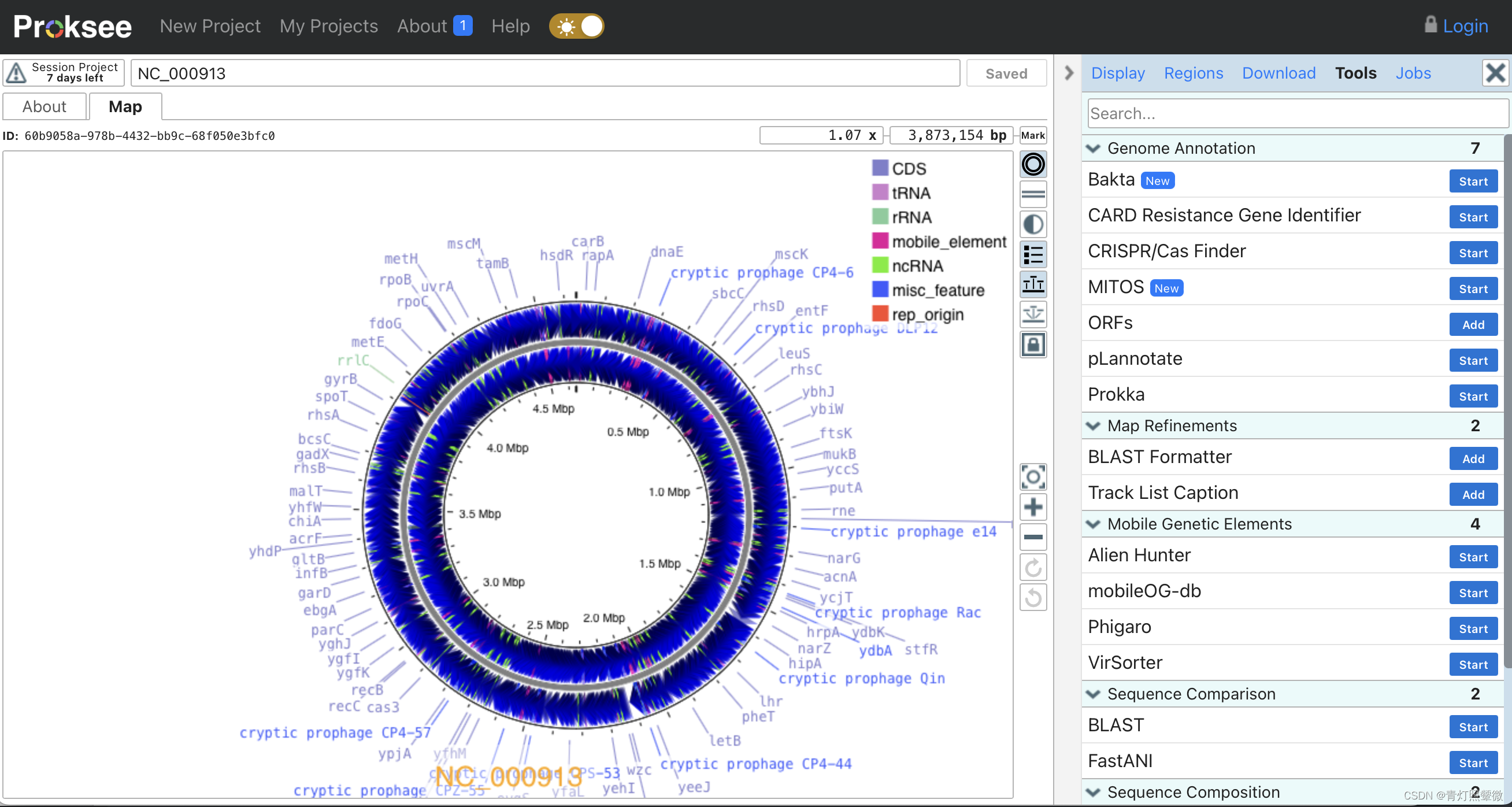
在线可视化工具Proksee
基因组组装、注释和可视化
https://proksee.ca
本文来自互联网用户投稿,该文观点仅代表作者本人,不代表本站立场。本站仅提供信息存储空间服务,不拥有所有权,不承担相关法律责任。 如若内容造成侵权/违法违规/事实不符,请联系我的编程经验分享网邮箱:chenni525@qq.com进行投诉反馈,一经查实,立即删除!