gseaplot3修改一下clusterProfiler默认绘图函数
发布时间:2024年01月11日
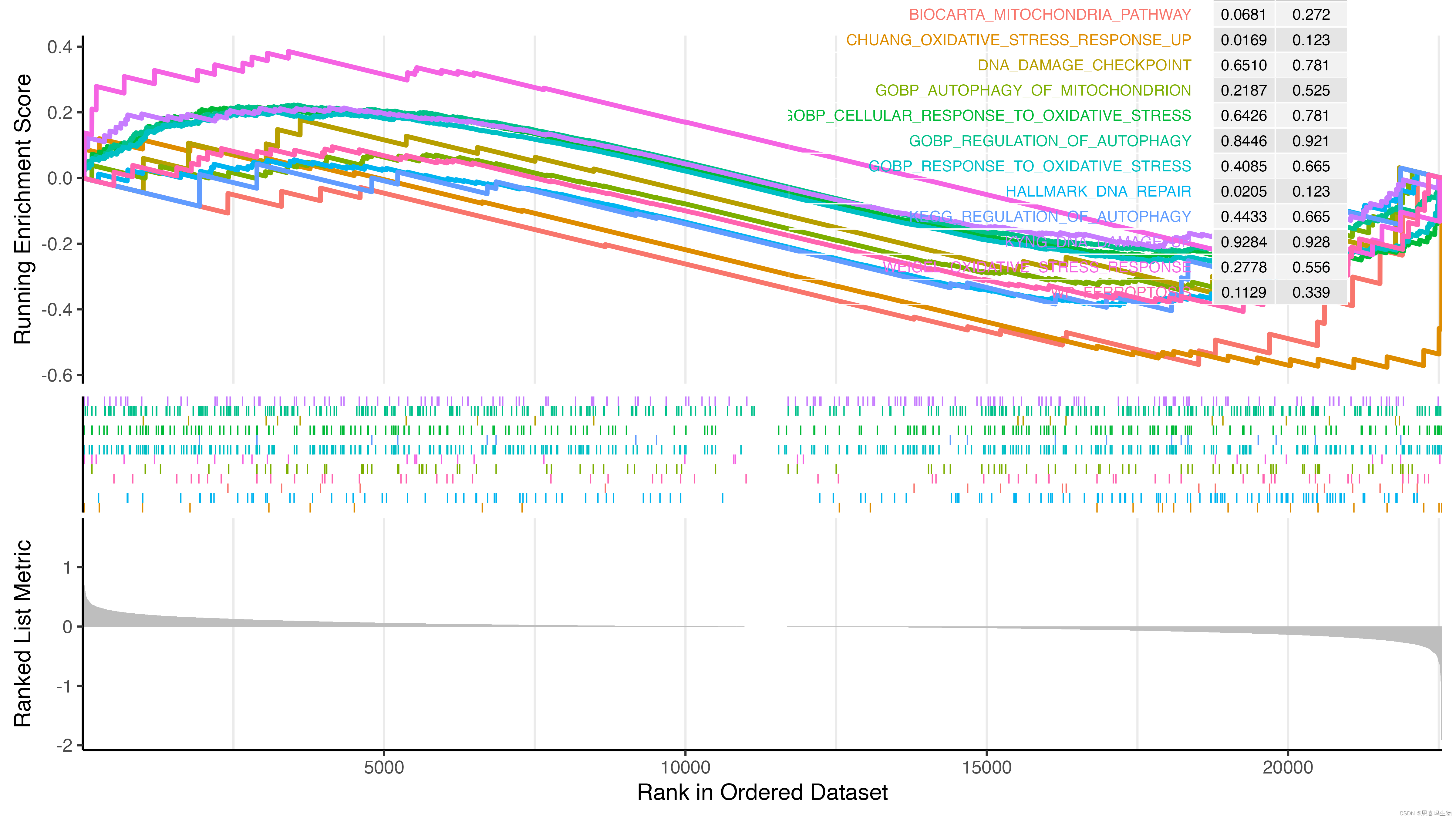
直接使用clusterProfiler::gseaplot2绘图会出现下边的结果,导致四周显示不全,线的粗细也没办法调整,因为返回的是一个aplot包中的gglist对象,没太多研究。
p1 <- clusterProfiler::gseaplot2(gsea_result, gsea_result$ID, pvalue_table = T, base_size = 18, ES_geom='line')

自定义gseaplot3函数增加了size参数调整线的粗细,也调整了margin四周边距,可以在下边gseaplot3函数的基础上继续调整,如果需要的话。
p2 <- gseaplot3(gsea_result, gsea_result$ID, pvalue_table = T, base_size = 18, ES_geom='line', size=1.8)
gseaplot3 <- function (x, geneSetID, title = "", color = "green", base_size = 15, size=1.8,
rel_heights = c(1.5, 0.5, 1), subplots = 1:3, pvalue_table = FALSE,
ES_geom = "line")
{
library(grid)
library(DOSE)
gseaScores <- getFromNamespace("gseaScores", "DOSE")
ES_geom <- match.arg(ES_geom, c("line", "dot"))
geneList <- position <- NULL
if (length(geneSetID) == 1) {
gsdata <- gsInfo(x, geneSetID)
}
else {
gsdata <- do.call(rbind, lapply(geneSetID, gsInfo, object = x))
}
p <- ggplot(gsdata, aes_(x = ~x)) + xlab(NULL) + theme_classic(base_size) +
theme(panel.grid.major = element_line(colour = "grey92"),
panel.grid.minor = element_line(colour = "grey92"),
panel.grid.major.y = element_blank(), panel.grid.minor.y = element_blank()) +
scale_x_continuous(expand = c(0, 0))
if (ES_geom == "line") {
es_layer <- geom_line(aes_(y = ~runningScore, color = ~Description),
size = size)
}
else {
es_layer <- geom_point(aes_(y = ~runningScore, color = ~Description),
size = size, data = subset(gsdata, position == 1))
}
p.res <- p + es_layer + theme(legend.position = c(0.8, 0.8),
legend.title = element_blank(), legend.background = element_rect(fill = "transparent"))
p.res <- p.res + ylab("Running Enrichment Score") + theme(axis.text.x = element_blank(),
axis.ticks.x = element_blank(), axis.line.x = element_blank(),
plot.margin = margin(t = 0.2, r = 0.2, b = 0, l = 0.2,
unit = "cm"))
i <- 0
for (term in unique(gsdata$Description)) {
idx <- which(gsdata$ymin != 0 & gsdata$Description ==
term)
gsdata[idx, "ymin"] <- i
gsdata[idx, "ymax"] <- i + 1
i <- i + 1
}
p2 <- ggplot(gsdata, aes_(x = ~x)) + geom_linerange(aes_(ymin = ~ymin,
ymax = ~ymax, color = ~Description)) + xlab(NULL) + ylab(NULL) +
theme_classic(base_size) + theme(legend.position = "none",
plot.margin = margin(t = -0.1, b = 0, unit = "cm"), axis.ticks = element_blank(),
axis.text = element_blank(), axis.line.x = element_blank()) +
scale_x_continuous(expand = c(0, 0)) + scale_y_continuous(expand = c(0,
0))
if (length(geneSetID) == 1) {
v <- seq(1, sum(gsdata$position), length.out = 9)
inv <- findInterval(rev(cumsum(gsdata$position)), v)
if (min(inv) == 0)
inv <- inv + 1
col <- c(rev(brewer.pal(5, "Blues")), brewer.pal(5, "Reds"))
ymin <- min(p2$data$ymin)
yy <- max(p2$data$ymax - p2$data$ymin) * 0.3
xmin <- which(!duplicated(inv))
xmax <- xmin + as.numeric(table(inv)[as.character(unique(inv))])
d <- data.frame(ymin = ymin, ymax = yy, xmin = xmin,
xmax = xmax, col = col[unique(inv)])
p2 <- p2 + geom_rect(aes_(xmin = ~xmin, xmax = ~xmax,
ymin = ~ymin, ymax = ~ymax, fill = ~I(col)), data = d,
alpha = 0.9, inherit.aes = FALSE)
}
df2 <- p$data
df2$y <- p$data$geneList[df2$x]
p.pos <- p + geom_segment(data = df2, aes_(x = ~x, xend = ~x,
y = ~y, yend = 0), color = "grey")
p.pos <- p.pos + ylab("Ranked List Metric") + xlab("Rank in Ordered Dataset") +
theme(plot.margin = margin(t = -0.1, r = 0.2, b = 0.2,
l = 0.2, unit = "cm"))
if (!is.null(title) && !is.na(title) && title != "")
p.res <- p.res + ggtitle(title)
if (length(color) == length(geneSetID)) {
p.res <- p.res + scale_color_manual(values = color)
if (length(color) == 1) {
p.res <- p.res + theme(legend.position = "none")
p2 <- p2 + scale_color_manual(values = "black")
}
else {
p2 <- p2 + scale_color_manual(values = color) + theme(legend.position = "none",plot.margin=margin(t = 0.1, r = 0.2, b = 0.1, l = 0.2, unit = "cm"))
}
}
if (pvalue_table) {
pd <- x[geneSetID, c("Description", "pvalue", "p.adjust")]
rownames(pd) <- pd$Description
pd <- pd[, -1]
for (i in seq_len(ncol(pd))) {
pd[, i] <- format(pd[, i], digits = 3)
}
tp <- tableGrob2(pd, p.res)
p.res <- p.res + theme(legend.position = "none",plot.margin=margin(t = 0.8, r = 0.2, b = 0.2, l = 0.2,unit = "cm")) + annotation_custom(tp,
xmin = quantile(p.res$data$x, 0.5), xmax = quantile(p.res$data$x,
0.95), ymin = quantile(p.res$data$runningScore,
0.75), ymax = quantile(p.res$data$runningScore,
0.9))
}
plotlist <- list(p.res, p2, p.pos)[subplots]
n <- length(plotlist)
plotlist[[n]] <- plotlist[[n]] + theme(axis.line.x = element_line(),
axis.ticks.x = element_line(), axis.text.x = element_text())
if (length(subplots) == 1)
return(plotlist[[1]] + theme(plot.margin = margin(t = 0.2,
r = 0.2, b = 0.2, l = 0.2, unit = "cm")))
if (length(rel_heights) > length(subplots))
rel_heights <- rel_heights[subplots]
aplot::gglist(gglist = plotlist, ncol = 1, heights = rel_heights)
}
gsInfo <- function(object, geneSetID) {
geneList <- object@geneList
if (is.numeric(geneSetID))
geneSetID <- object@result[geneSetID, "ID"]
geneSet <- object@geneSets[[geneSetID]]
exponent <- object@params[["exponent"]]
df <- gseaScores(geneList, geneSet, exponent, fortify=TRUE)
df$ymin <- 0
df$ymax <- 0
pos <- df$position == 1
h <- diff(range(df$runningScore))/20
df$ymin[pos] <- -h
df$ymax[pos] <- h
df$geneList <- geneList
df$Description <- object@result[geneSetID, "Description"]
return(df)
}
tableGrob2 <- function(d, p = NULL) {
# has_package("gridExtra")
d <- d[order(rownames(d)),]
tp <- gridExtra::tableGrob(d)
if (is.null(p)) {
return(tp)
}
# Fix bug: The 'group' order of lines and dots/path is different
p_data <- ggplot_build(p)$data[[1]]
# pcol <- unique(ggplot_build(p)$data[[1]][["colour"]])
p_data <- p_data[order(p_data[["group"]]), ]
pcol <- unique(p_data[["colour"]])
## This is fine too
## pcol <- unique(p_data[["colour"]])[unique(p_data[["group"]])]
j <- which(tp$layout$name == "rowhead-fg")
for (i in seq_along(pcol)) {
tp$grobs[j][[i+1]][["gp"]] <- gpar(col = pcol[i])
}
return(tp)
}

文章来源:https://blog.csdn.net/weixin_44493991/article/details/135526390
本文来自互联网用户投稿,该文观点仅代表作者本人,不代表本站立场。本站仅提供信息存储空间服务,不拥有所有权,不承担相关法律责任。 如若内容造成侵权/违法违规/事实不符,请联系我的编程经验分享网邮箱:chenni525@qq.com进行投诉反馈,一经查实,立即删除!
本文来自互联网用户投稿,该文观点仅代表作者本人,不代表本站立场。本站仅提供信息存储空间服务,不拥有所有权,不承担相关法律责任。 如若内容造成侵权/违法违规/事实不符,请联系我的编程经验分享网邮箱:chenni525@qq.com进行投诉反馈,一经查实,立即删除!
最新文章
- Python教程
- 深入理解 MySQL 中的 HAVING 关键字和聚合函数
- Qt之QChar编码(1)
- MyBatis入门基础篇
- 用Python脚本实现FFmpeg批量转换
- 锐捷交换机配置 VRRP(网关冗余协议)
- PB 按Excel动态创建对应字段
- MySQL函数—字符串函数
- Scrapy爬虫在新闻数据提取中的应用
- 基于NXP I.MX8 + Codesys的工业软PLC解决方案
- 【设计模式】中介模式
- LED恒流驱动芯片SM2188EN:满足LED灯具出口欧盟所需的ERP能效认证标准和要求
- STM32-I2C通讯-AHT20温湿度检测
- 16-网络安全框架及模型-BiBa完整性模型
- 链接器--静态链接学习笔记二-----变量与内存地址是如何映射的?