生信软件10 - DNA/RNA/蛋白多序列比对图R包ggmsa
发布时间:2023年12月26日
R包安装
BiocManager::install("ggplot2")
BiocManager::install("ggmsa")
DNA多序列比对图
# 导入内置核酸序列
nt_sequences <- system.file("extdata", "LeaderRepeat_All.fa", package = "ggmsa")
ggmsa(nt_sequences, color="Chemistry_AA", font = "DroidSansMono", char_width = 0.5, seq_name = TRUE)

RNA多序列比对图
miRNA_sequences <- system.file("extdata", "seedSample.fa", package = "ggmsa")
ggmsa(miRNA_sequences, font = 'DroidSansMono',color = "Chemistry_NT", none_bg = TRUE) +
geom_seed(seed = "GAGGUAG", star = FALSE)ggmsa(miRNA_sequences, font = 'DroidSansMono',color = "Chemistry_NT") +
geom_seed(seed = "GAGGUAG", star = TRUE)

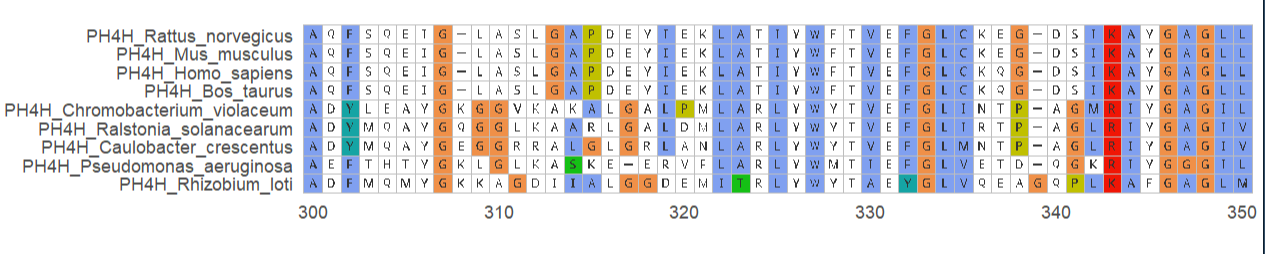
蛋白质多序列比对图
protein_sequences <- system.file("extdata", "sample.fasta", package = "ggmsa")
ggmsa(protein_sequences, 221, 280, seq_name = TRUE, char_width = 0.5) +
geom_seqlogo(color = "Chemistry_AA") + geom_msaBar()
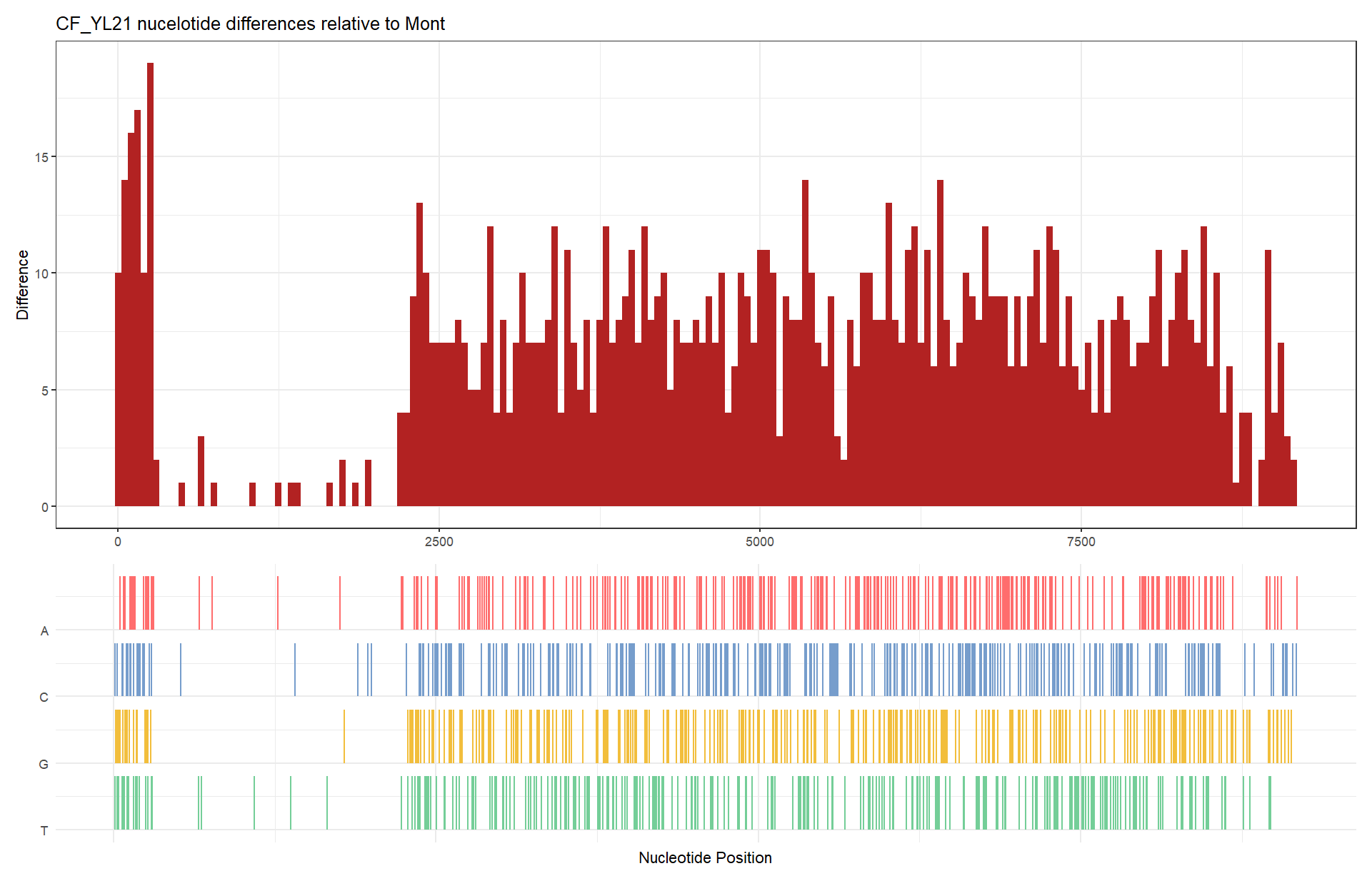
基因组变异图
fas <- list.files(system.file("extdata", "GVariation", package="ggmsa"), pattern="fas", full.names=TRUE)
x <- seqdiff(fas[1], reference=1)
plot(
x,
width = 50,
title = "auto",
xlab = "Nucleotide Position",
by = "bar",
fill = "firebrick",
colors = c(A = "#ff6d6d", C = "#769dcc", G = "#f2be3c", T = "#74ce98"),
xlim = NULL
)
生信软件文章推荐
生信软件1 - 测序下机文件比对结果可视化工具 visNano
生信软件3 - mapping比对bam文件质量评估工具 qualimap
生信软件4 - 拷贝数变异CNV分析软件 WisecondorX
生信软件7 - 多线程并行运行Linux效率工具Parallel
生信软件8 - bedtools进行窗口划分、窗口GC含量、窗口测序深度和窗口SNP统计
生信软件9 - 多公共数据库数据下载软件Kingfisher
更多内容请关注公众号【生信与基因组学】,定期更新生信算法和编程、基因组学、统计学、分子生物学、临床检测和深度学习等内容。
文章来源:https://blog.csdn.net/LittleComputerRobot/article/details/135231288
本文来自互联网用户投稿,该文观点仅代表作者本人,不代表本站立场。本站仅提供信息存储空间服务,不拥有所有权,不承担相关法律责任。 如若内容造成侵权/违法违规/事实不符,请联系我的编程经验分享网邮箱:chenni525@qq.com进行投诉反馈,一经查实,立即删除!
本文来自互联网用户投稿,该文观点仅代表作者本人,不代表本站立场。本站仅提供信息存储空间服务,不拥有所有权,不承担相关法律责任。 如若内容造成侵权/违法违规/事实不符,请联系我的编程经验分享网邮箱:chenni525@qq.com进行投诉反馈,一经查实,立即删除!
最新文章
- Python教程
- 深入理解 MySQL 中的 HAVING 关键字和聚合函数
- Qt之QChar编码(1)
- MyBatis入门基础篇
- 用Python脚本实现FFmpeg批量转换
- realsense t265参数查询与释义
- ISA Server2006部署RuoYi无法登录的问题
- c# OpenCV 图像裁剪、调整大小、旋转、透视(三)
- CSS背景属性和显示模式
- Windows内网渗透篇-后门持久化姿势总结
- 如何在苹果手机上进行文件管理
- JSON&yaml和Properties
- MySQL乐观锁与悲观锁
- PD SINK协议芯片系列产品介绍对比-ECP5701、FS312A、CH221K、HUSB238、AS225KL
- 连接打印机显示“0x0000011b”错误代码,亲测有效的解决办法