OpenCV实战 -- 维生素药片的检测记数
发布时间:2023年12月31日
文章目录
读取图片
形态学处理
二值化
提取轮廓
获取轮廓索引,并筛选所需要的轮廓
画出轮廓,显示计数
检测记数
原图-》灰度化-》阈值分割-》形态学变换-》距离变换-》轮廓查找
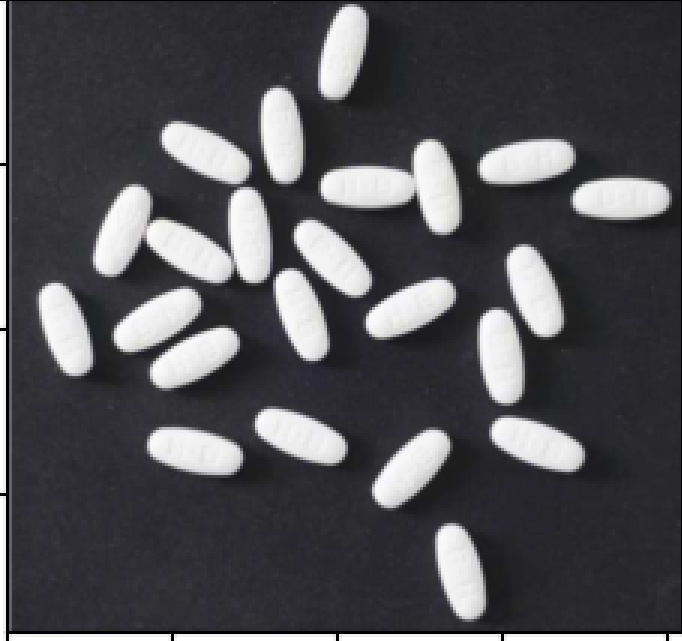
原图
import cv2 as cv
import matplotlib.pyplot as plt
image = cv.imread('img/img.png')
gray_image = cv.cvtColor(image, cv.COLOR_BGR2GRAY)
ret, binary = cv.threshold(gray_image, 127, 255, cv.THRESH_BINARY)
# 寻找轮廓
contours, hierarchy = cv.findContours(binary, cv.RETR_TREE, cv.CHAIN_APPROX_SIMPLE)
# 在原始图像的副本上绘制轮廓并标注序号
image_with_contours = image.copy()
for i, contour in enumerate(contours):
cv.drawContours(image_with_contours, [contour], -1, (122, 55, 215), 2)
# 标注轮廓序号
cv.putText(image_with_contours, str(i+1), tuple(contour[0][0]), cv.FONT_HERSHEY_SIMPLEX, 0.5, (0, 255, 0), 2)
# 使用 matplotlib 显示结果
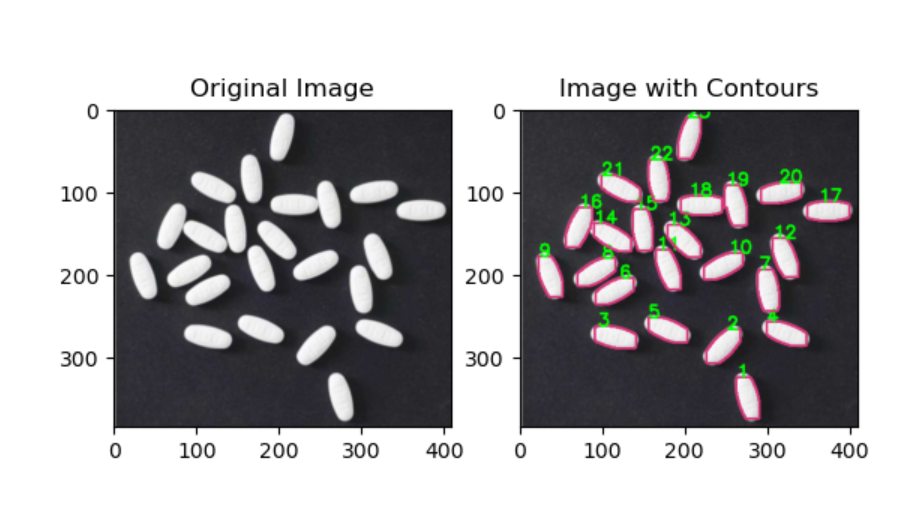
plt.subplot(121), plt.imshow(cv.cvtColor(image, cv.COLOR_BGR2RGB)), plt.title('Original Image')
plt.subplot(122), plt.imshow(cv.cvtColor(image_with_contours, cv.COLOR_BGR2RGB)), plt.title('Image with Contours')
plt.show()
print (len(contours))
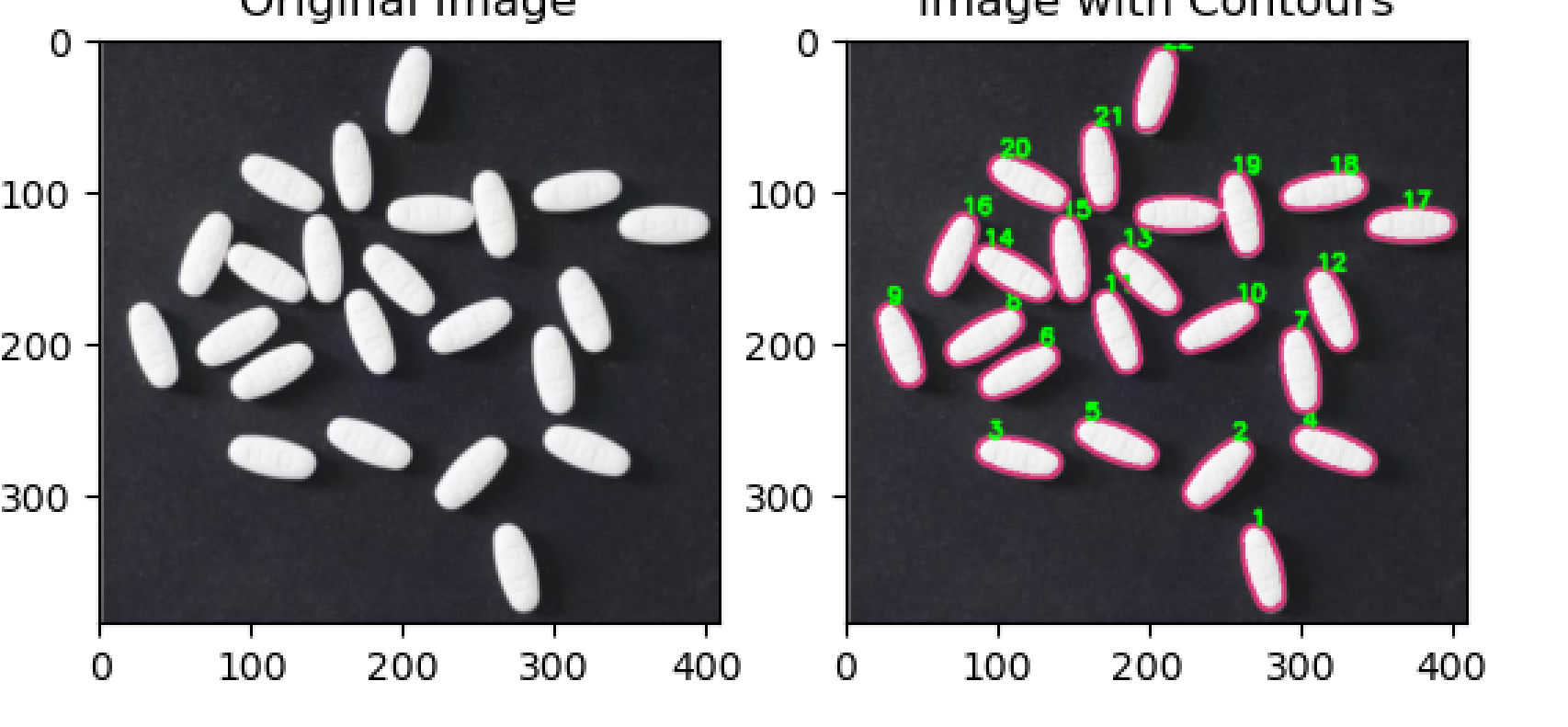
经过操作
发现其具有粘连性,所以阈值分割、形态学变换等图像处理
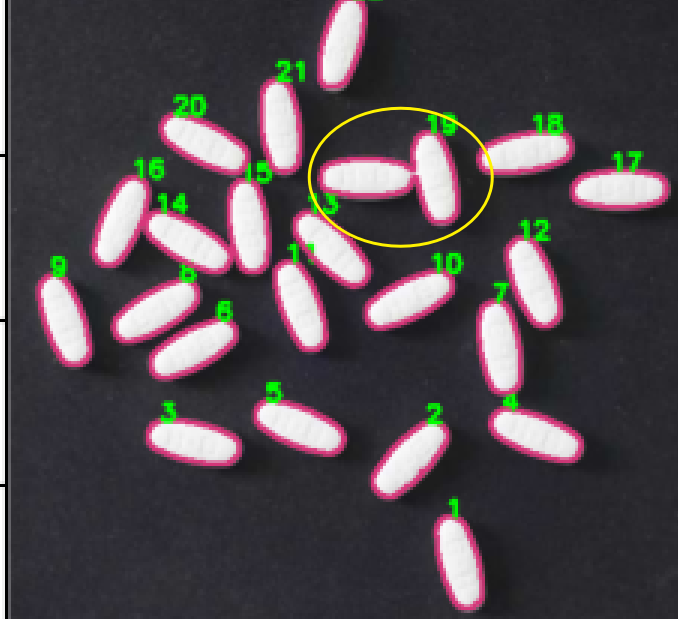
开始进行消除粘连性–形态学变换
import numpy as np
import cv2 as cv
import matplotlib.pyplot as plt
image = cv.imread('img/img.png')
gray_image= cv.cvtColor(image, cv.COLOR_BGR2GRAY)
kernel = np.ones((16, 16), np.uint8)
gray_image=cv.morphologyEx(gray_image, cv.MORPH_OPEN, kernel)
ret, binary = cv.threshold(gray_image, 127, 255, cv.THRESH_BINARY)
# 寻找轮廓
contours, hierarchy = cv.findContours(binary, cv.RETR_TREE, cv.CHAIN_APPROX_SIMPLE)
# 在原始图像的副本上绘制轮廓并标注序号
image_with_contours = image.copy()
for i, contour in enumerate(contours):
cv.drawContours(image_with_contours, [contour], -1, (122, 55, 215), 2)
# 标注轮廓序号
cv.putText(image_with_contours, str(i+1), tuple(contour[0][0]), cv.FONT_HERSHEY_SIMPLEX, 0.7, (0, 255, 0), 2)
# 使用 matplotlib 显示结果
plt.subplot(121), plt.imshow(cv.cvtColor(image, cv.COLOR_BGR2RGB)), plt.title('Original Image')
plt.subplot(122), plt.imshow(cv.cvtColor(image_with_contours, cv.COLOR_BGR2RGB)), plt.title('Image with Contours')
plt.show()
print (len(contours))
总结实现方法
1. 读取图片:
import cv2
# 读取图片
image = cv2.imread("path/to/your/image.png")
cv2.imshow("Original Image", image)
cv2.waitKey(0)
2. 形态学处理:
import cv2
import numpy as np
# 形态学处理
kernel = np.ones((16, 16), np.uint8)
morphology_result = cv2.morphologyEx(image, cv2.MORPH_OPEN, kernel)
cv2.imshow("Morphology Result", morphology_result)
cv2.waitKey(0)
3. 二值化:
import cv2
# 灰度转换
gray_image = cv2.cvtColor(morphology_result, cv2.COLOR_BGR2GRAY)
# 二值化
_, binary_image = cv2.threshold(gray_image, 100, 255, cv2.THRESH_OTSU)
cv2.imshow("Binary Image", binary_image)
cv2.waitKey(0)
4. 提取轮廓:
import cv2
# 寻找轮廓
contours, _ = cv2.findContours(binary_image, cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_NONE)
# 在原图上绘制轮廓
contour_image = image.copy()
cv2.drawContours(contour_image, contours, -1, (0, 255, 0), 2)
cv2.imshow("Contours", contour_image)
cv2.waitKey(0)
5. 轮廓筛选和计数:
import cv2
# 遍历轮廓
for i, contour in enumerate(contours):
area = cv2.contourArea(contour)
if area < 500:
continue
# 获取轮廓的位置
(x, y, w, h) = cv2.boundingRect(contour)
# 在原图上绘制矩形
cv2.rectangle(image, (x, y), (x + w, y + h), (0, 255, 0), 2)
# 在矩形位置写上计数
cv2.putText(image, str(i), (x, y), cv2.FONT_HERSHEY_COMPLEX, 1, (0, 0, 255), 2)
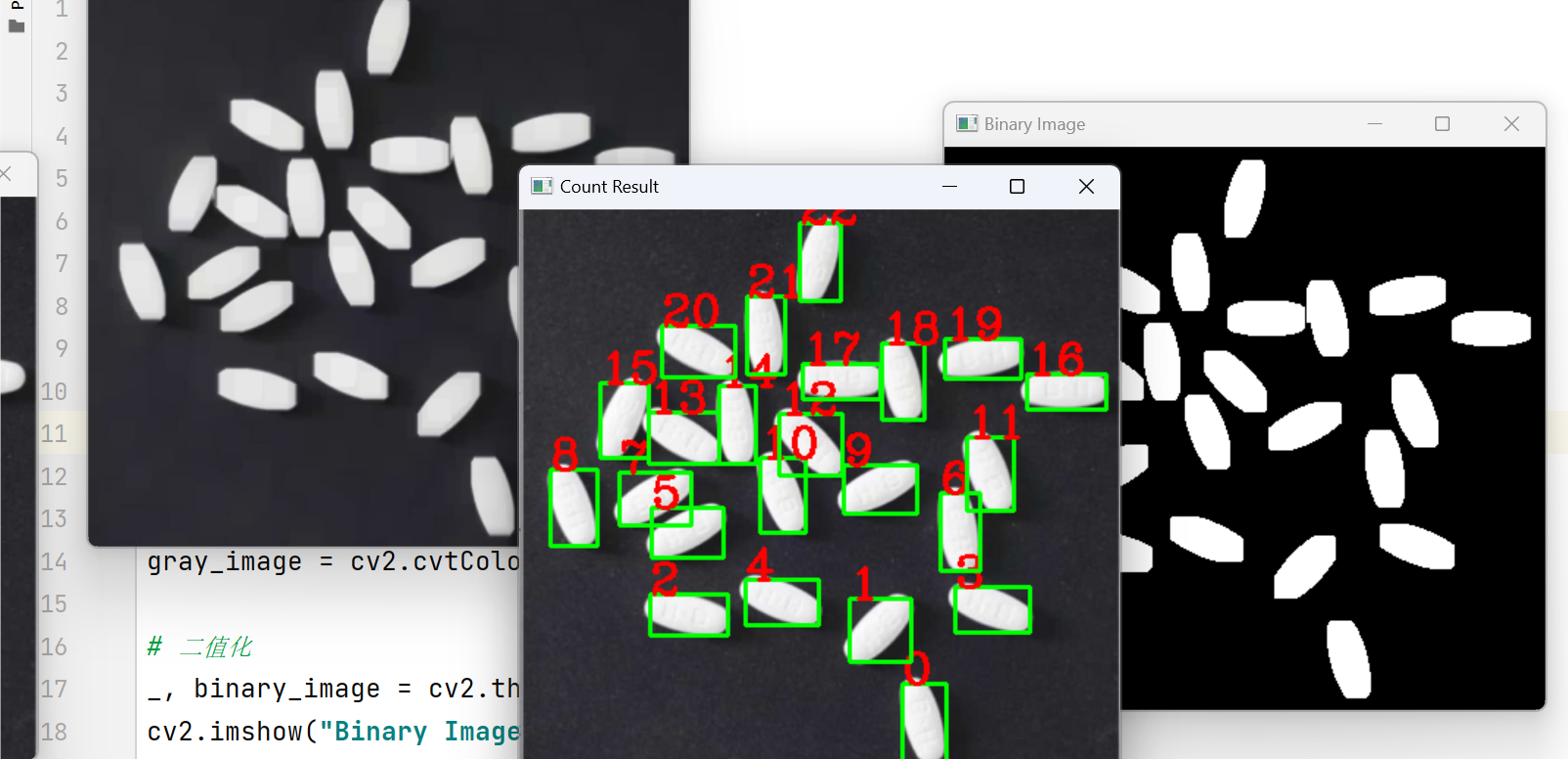
cv2.imshow("Count Result", image)
cv2.waitKey(0)
分水岭算法:
import cv2
import numpy as np
# 读取图片
image = cv2.imread("path/to/your/image.png")
cv2.imshow("Original Image", image)
# 形态学处理
kernel = np.ones((3, 3), np.uint8)
morphology_result = cv2.morphologyEx(image, cv2.MORPH_OPEN, kernel)
cv2.imshow("Morphology Result", morphology_result)
# 灰度转换
gray_image = cv2.cvtColor(morphology_result, cv2.COLOR_BGR2GRAY)
# 二值化
_, binary_image = cv2.threshold(gray_image, 100, 255, cv2.THRESH_OTSU)
cv2.imshow("Binary Image", binary_image)
# 寻找轮廓
contours, _ = cv2.findContours(binary_image, cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_NONE)
# 统计药片数量并标记轮廓
count = 0
for i, contour in enumerate(contours):
area = cv2.contourArea(contour)
if area < 500:
continue
# 获取轮廓的位置
(x, y, w, h) = cv2.boundingRect(contour)
# 在原图上绘制矩形
cv2.rectangle(image, (x, y), (x + w, y + h), (0, 255, 0), 2)
# 在矩形位置写上计数
cv2.putText(image, str(count), (x, y), cv2.FONT_HERSHEY_COMPLEX, 1, (0, 0, 255), 2)
count += 1
cv2.imshow("Count Result", image)
print("药片检测个数:", count)
cv2.waitKey(0)
cv2.destroyAllWindows()
逐行解释
当然,让我们逐行解释上述代码:
import cv2
import numpy as np
# 读取图片
image = cv2.imread("path/to/your/image.png")
cv2.imshow("Original Image", image)
- 导入OpenCV库和NumPy库。
- 读取图片并显示原始图像。
# 形态学处理
kernel = np.ones((3, 3), np.uint8)
morphology_result = cv2.morphologyEx(image, cv2.MORPH_OPEN, kernel)
cv2.imshow("Morphology Result", morphology_result)
- 定义一个3x3的矩形内核(kernel)。
- 对原始图像进行形态学开运算,去除小的噪点和不重要的细节。
- 显示形态学处理后的图像。
# 灰度转换
gray_image = cv2.cvtColor(morphology_result, cv2.COLOR_BGR2GRAY)
- 将形态学处理后的图像转换为灰度图。
# 二值化
_, binary_image = cv2.threshold(gray_image, 100, 255, cv2.THRESH_OTSU)
cv2.imshow("Binary Image", binary_image)
- 对灰度图进行自适应阈值二值化,使用OTSU算法。
- 显示二值化后的图像。
# 寻找轮廓
contours, _ = cv2.findContours(binary_image, cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_NONE)
- 寻找二值化后图像中的外部轮廓。
# 统计药片数量并标记轮廓
count = 0
for i, contour in enumerate(contours):
area = cv2.contourArea(contour)
if area < 500:
continue
# 获取轮廓的位置
(x, y, w, h) = cv2.boundingRect(contour)
# 在原图上绘制矩形
cv2.rectangle(image, (x, y), (x + w, y + h), (0, 255, 0), 2)
# 在矩形位置写上计数
cv2.putText(image, str(count), (x, y), cv2.FONT_HERSHEY_COMPLEX, 1, (0, 0, 255), 2)
count += 1
cv2.imshow("Count Result", image)
print("药片检测个数:", count)
- 初始化药片计数为0。
- 遍历所有找到的轮廓。
- 如果轮廓的面积小于500,则跳过。
- 获取轮廓的位置信息(矩形边界框)。
- 在原图上绘制矩形,标记检测到的药片。
- 在矩形位置写上计数。
- 计数加1。
- 显示标记了计数的结果图像,并输出药片检测个数。
cv2.waitKey(0)
cv2.destroyAllWindows()
- 等待用户按下任意按键,然后关闭所有打开的窗口。
在基于距离变换的分水岭算法中,二值化操作是为了得到sure_fg(肯定是前景的区域),以便将其用作分水岭算法的标记点。这个过程涉及以下几步:
-
距离变换: 通过距离变换,我们得到了一个灰度图,其中像素值表示每个像素到最近的零像素点的距离。这个距离图范围是浮点数,通常需要进行归一化。
dist_transform = cv2.distanceTransform(binary_image, cv2.DIST_L2, 3) -
归一化: 将距离变换后的图像进行归一化,使其范围在0到1之间。
normalized_distance = cv2.normalize(dist_transform, 0, 1, cv2.NORM_MINMAX) -
再次二值化: 对归一化后的图像进行二值化,以获取肯定是前景的区域。这是通过设置一个阈值,将距离较大的区域认定为前景。
_, sure_fg = cv2.threshold(normalized_distance, 0.4, 1, cv2.THRESH_BINARY)
这样,sure_fg 中的像素值为 1 的区域就被认为是明确的前景区域,而不是可能的边界区域。这种区域将被用作分水岭算法的种子点。
文章来源:https://blog.csdn.net/m0_74154295/article/details/134621904
本文来自互联网用户投稿,该文观点仅代表作者本人,不代表本站立场。本站仅提供信息存储空间服务,不拥有所有权,不承担相关法律责任。 如若内容造成侵权/违法违规/事实不符,请联系我的编程经验分享网邮箱:chenni525@qq.com进行投诉反馈,一经查实,立即删除!
本文来自互联网用户投稿,该文观点仅代表作者本人,不代表本站立场。本站仅提供信息存储空间服务,不拥有所有权,不承担相关法律责任。 如若内容造成侵权/违法违规/事实不符,请联系我的编程经验分享网邮箱:chenni525@qq.com进行投诉反馈,一经查实,立即删除!
最新文章
- Python教程
- 深入理解 MySQL 中的 HAVING 关键字和聚合函数
- Qt之QChar编码(1)
- MyBatis入门基础篇
- 用Python脚本实现FFmpeg批量转换
- uniapp小程序当页面内容超出时显示滚动条,不超出时不显示---样式自定义
- 服务器配置优化句柄数量
- Springboot3新特性:GraalVM Native Image Support和虚拟线程(从入门到精通)
- STM32——串口通信应用篇
- 组件生产ERP软件是什么?组件生产ERP系统怎么样
- 【Docker】未来已来 | Docker技术在云计算、边缘计算领域的应用前景
- 代码随想录 Leetcode203. 移除链表元素
- FPGA_ZYNQ (PS端)开发流程(Xilinx软件工具介绍)
- 五、交换机基础配置实验
- 三本光电从颓废到武汉年薪30w的本科经历经验与浅谈(毕业工作一年的嵌入式软件工程师经验分享)