基于深度学习神经网络cnn的柑橘病虫害识别系统源码
发布时间:2024年01月10日
?第一步:准备数据
5种柑橘病虫数据:Fruit-anthrax,Fruit-ulcer,leaf_thyroid,Leaf-anthrax和Leaf-ulcer,总共有3030张图片,每个文件夹单独放一种数据

第二步:搭建模型
本文用的是resnet50模型,其网络结构如下:
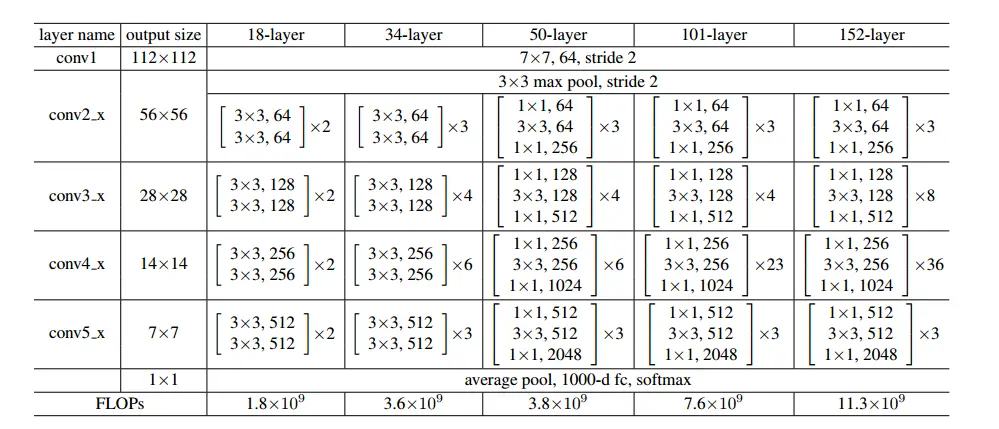
模型代码如下:
def resnet_50(IMG_SHAPE=(224, 224, 3), class_num=5):
inpt =Input(shape=IMG_SHAPE)
x = ZeroPadding2D((3, 3))(inpt)
x = Conv2d_BN(x, nb_filter=64, kernel_size=(7, 7), strides=(2, 2), padding='valid')
x = MaxPooling2D(pool_size=(3, 3), strides=(2, 2), padding='same')(x)
#conv2_x
x = bottleneck_Block(x, nb_filters=[64,64,256],strides=(1,1),with_conv_shortcut=True)
x = bottleneck_Block(x, nb_filters=[64,64,256])
x = bottleneck_Block(x, nb_filters=[64,64,256])
#conv3_x
x = bottleneck_Block(x, nb_filters=[128, 128, 512],strides=(2,2),with_conv_shortcut=True)
x = bottleneck_Block(x, nb_filters=[128, 128, 512])
x = bottleneck_Block(x, nb_filters=[128, 128, 512])
x = bottleneck_Block(x, nb_filters=[128, 128, 512])
#conv4_x
x = bottleneck_Block(x, nb_filters=[256, 256, 1024],strides=(2,2),with_conv_shortcut=True)
x = bottleneck_Block(x, nb_filters=[256, 256, 1024])
x = bottleneck_Block(x, nb_filters=[256, 256, 1024])
x = bottleneck_Block(x, nb_filters=[256, 256, 1024])
x = bottleneck_Block(x, nb_filters=[256, 256, 1024])
x = bottleneck_Block(x, nb_filters=[256, 256, 1024])
#conv5_x
x = bottleneck_Block(x, nb_filters=[512, 512, 2048], strides=(2, 2), with_conv_shortcut=True)
x = bottleneck_Block(x, nb_filters=[512, 512, 2048])
x = bottleneck_Block(x, nb_filters=[512, 512, 2048])
x = AveragePooling2D(pool_size=(7, 7))(x)
x = Flatten()(x)
x = Dense(class_num, activation='softmax')(x)
model = Model(inputs=inpt, outputs=x)
# 输出模型信息
model.summary()
# 指明模型的训练参数,优化器为sgd优化器,损失函数为交叉熵损失函数
model.compile(optimizer='sgd', loss='categorical_crossentropy', metrics=['accuracy'])
# 返回模型
return model第三步:训练过程中的部分中间结果
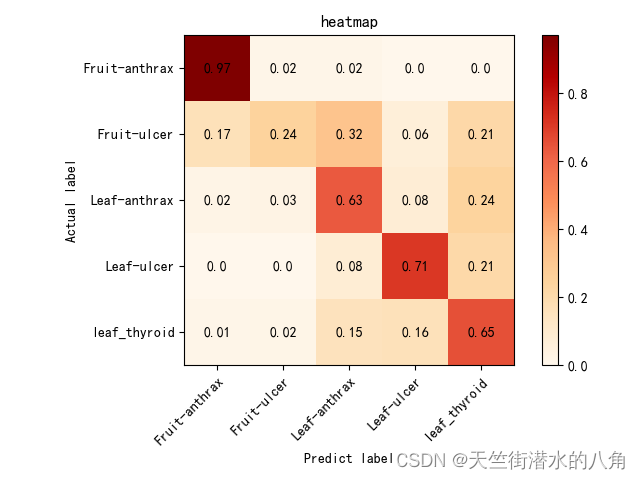
混淆矩阵
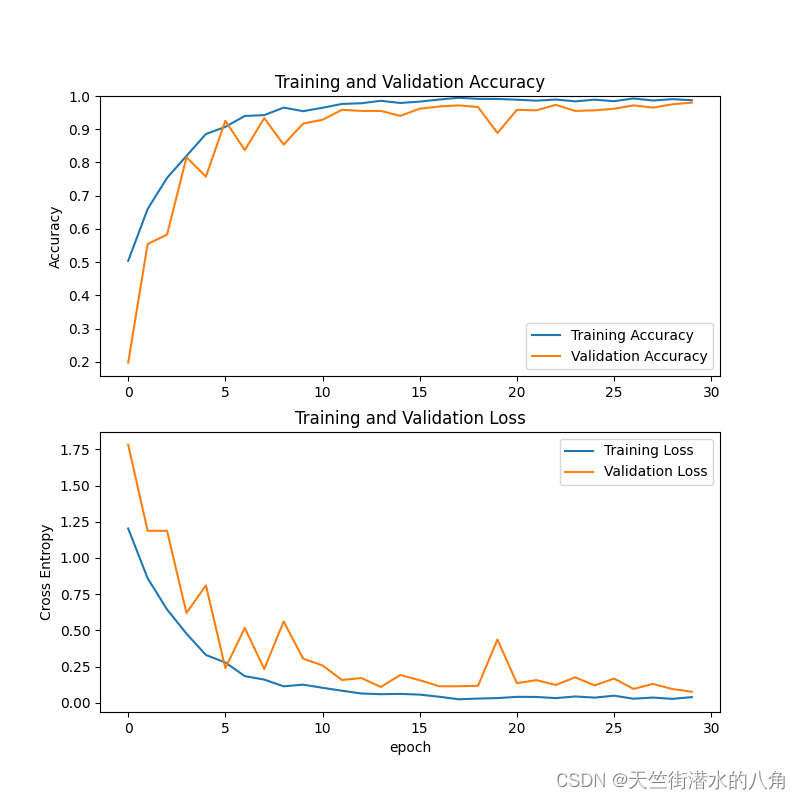
正确率和loss变化:
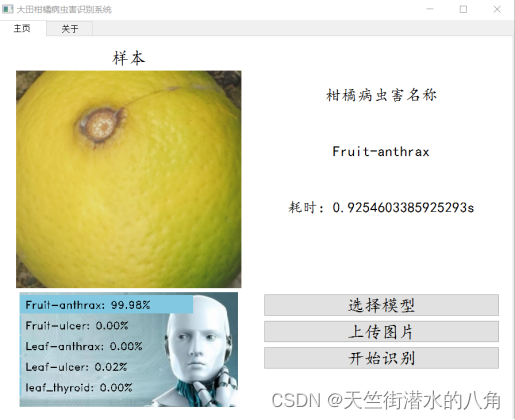
第四步:搭建GUI界面
第六步:整个工程的内容
提供整套,提供测试数据,提供GUI界面代码,主要使用方法可以参考里面的“文档说明_必看.docx”
代码的下载路径(新窗口打开链接):基于深度学习神经网络cnn的柑橘病虫害识别系统源码

有问题可以私信或者留言,有问必答
文章来源:https://blog.csdn.net/m0_59023219/article/details/135485526
本文来自互联网用户投稿,该文观点仅代表作者本人,不代表本站立场。本站仅提供信息存储空间服务,不拥有所有权,不承担相关法律责任。 如若内容造成侵权/违法违规/事实不符,请联系我的编程经验分享网邮箱:chenni525@qq.com进行投诉反馈,一经查实,立即删除!
本文来自互联网用户投稿,该文观点仅代表作者本人,不代表本站立场。本站仅提供信息存储空间服务,不拥有所有权,不承担相关法律责任。 如若内容造成侵权/违法违规/事实不符,请联系我的编程经验分享网邮箱:chenni525@qq.com进行投诉反馈,一经查实,立即删除!
最新文章
- Python教程
- 深入理解 MySQL 中的 HAVING 关键字和聚合函数
- Qt之QChar编码(1)
- MyBatis入门基础篇
- 用Python脚本实现FFmpeg批量转换
- SpringBoot 集成 Nacos,附带Nacos的安装
- AI大模型引领未来智慧科研暨丨ChatGPT在地学、GIS、气象、农业、生态、环境等领域中的高级应用
- 外包干了2个月,技术退步明显了...
- cargo run 报错error: linking with `cc` failed: exit status: 1
- 22k+stars centos轻松搭建网盘的神器
- java distinct 无法为泛型去重
- pc端微信&QQ使用代理解决方案
- MongoDB
- 微信小程序---全局数据共享
- 力扣5. 最长回文子串